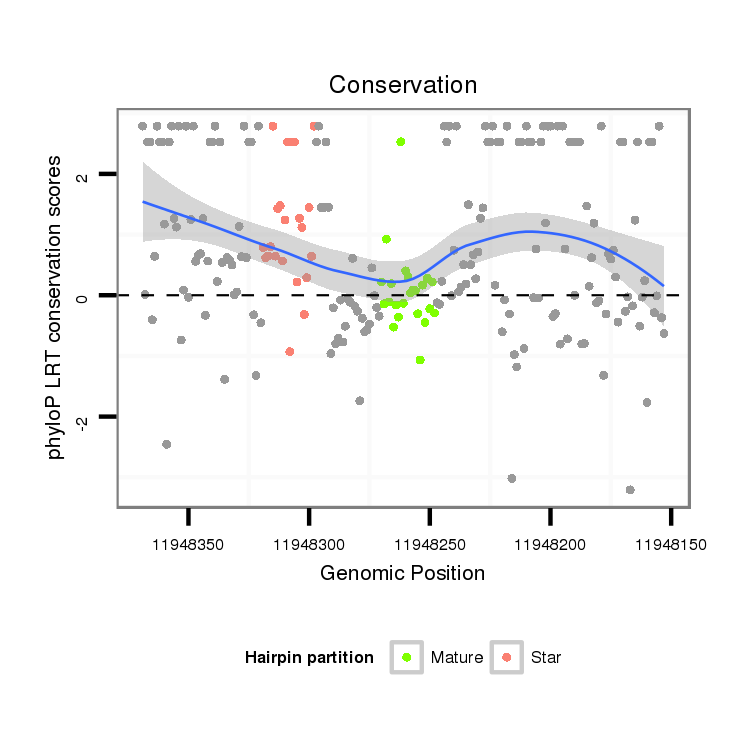
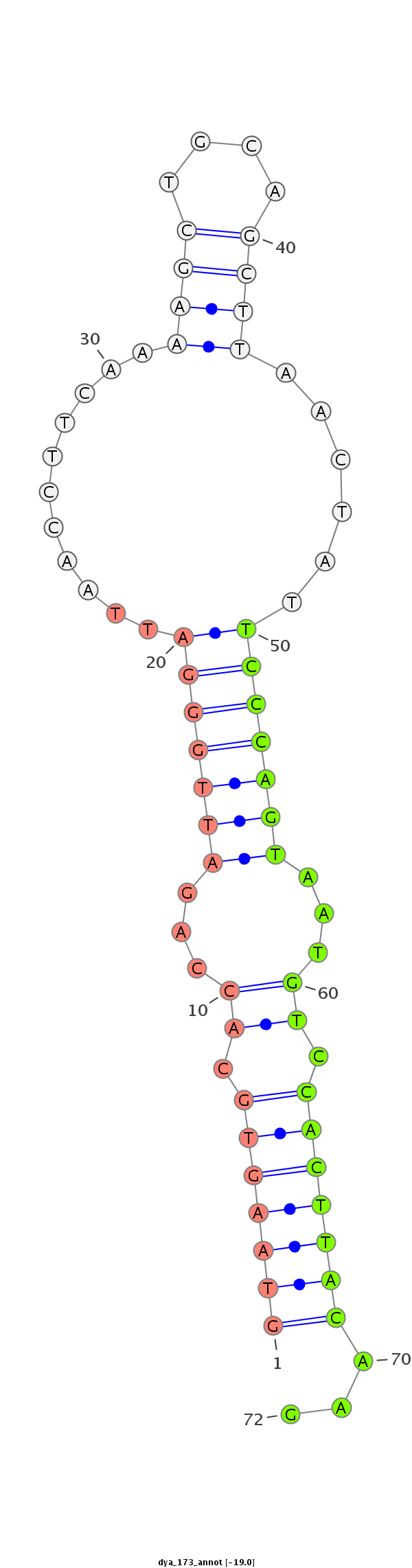
ID:dya_173 |
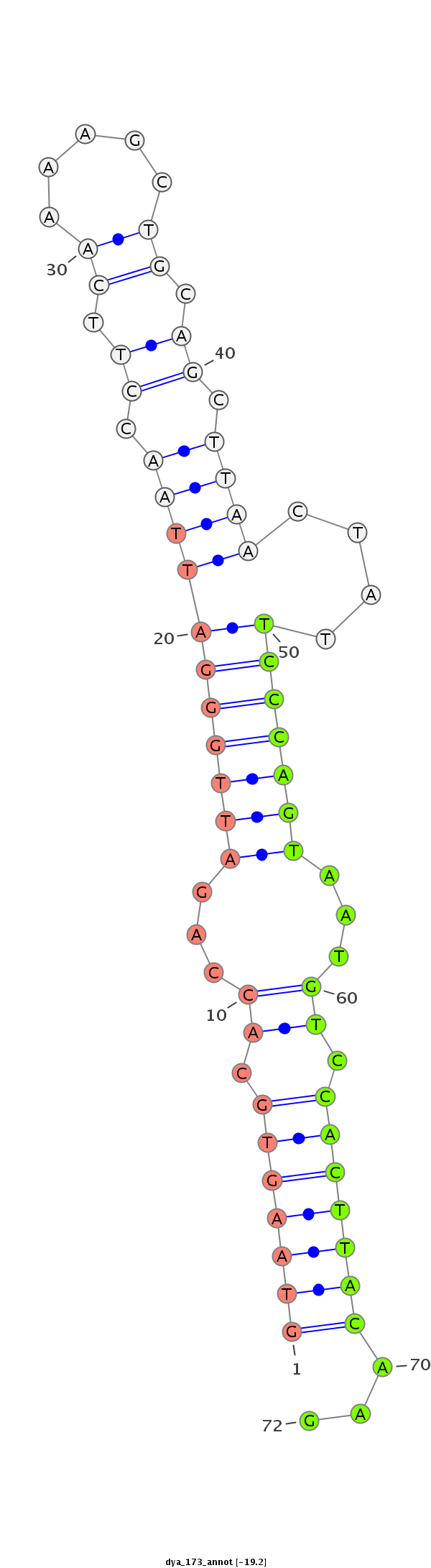
Coordinate:3R:11948248-11948319 - |
Confidence:Known Ortholog |
Class:Mirtron |
Genomic Locale:5pUTR |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -19.0 | -19.0 | -19.0 |
 |
 |
 |
utr5 [utr5_plus_10969]; CDS [Dyak\GE24582-cds]; intron [Dyak\GE24582-in]; CDS [Dyak\GE24582-cds]
No Repeatable elements found
|
CCTGCGGGAGGCGGGCGACCTGCCACTGTGCATCTTCCGCCCGGCGATCAGTAAGTGCACCAGATTGGGATTAACCTTCAAAAGCTGCAGCTTAACTATTCCCAGTAATGTCCACTTACAAGGAGCCCCTGGATGGCTGGGTGGACAACCTATTCGGACCATTGGCCCTCTG
**************************************************(((((((.((...(((((((((((.((.((.....)).)).))))....)))))))...)).)))))))...************************************************** |
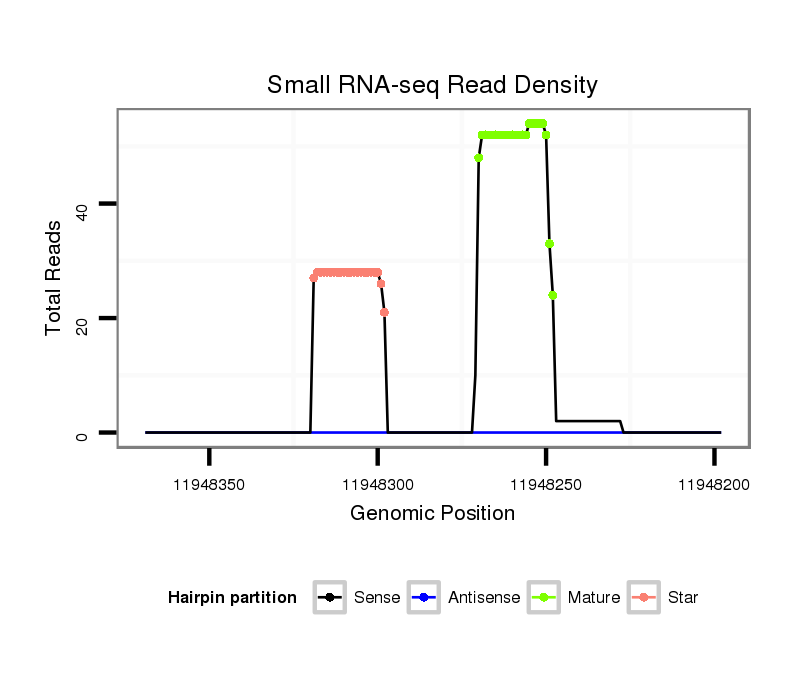
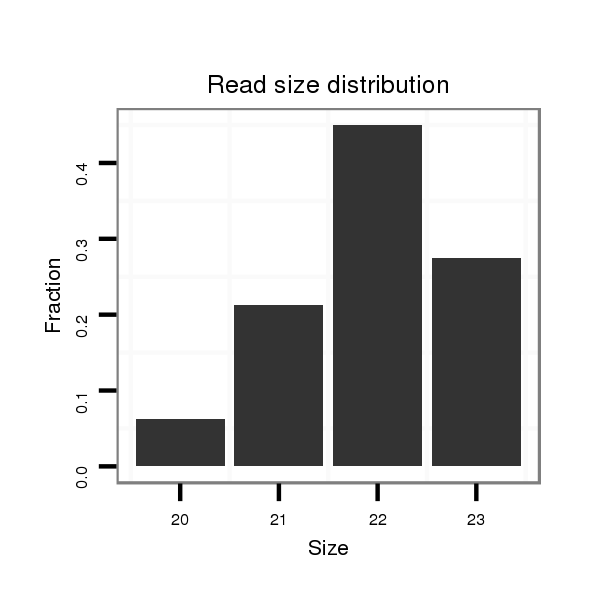
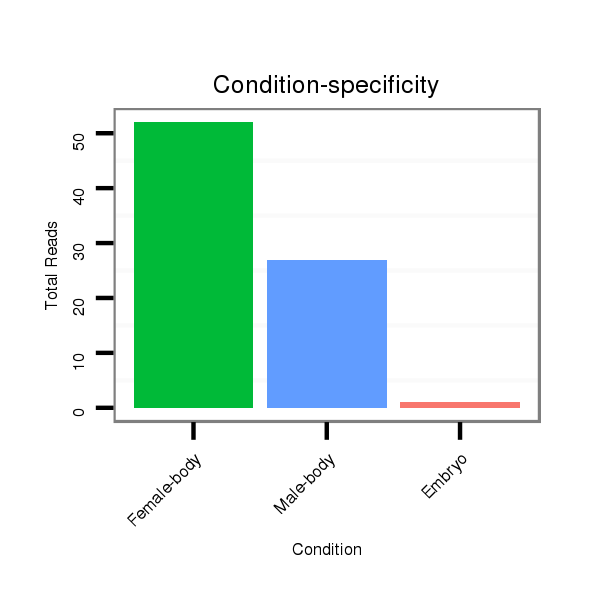
Read size | # Mismatch | Hit Count | Total Norm | Total | M043 female body |
V120 male body |
M056 embryo |
V058 head |
|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................TCCCAGTAATGTCCACTTACAAG.................................................. | 23 | 0 | 1 | 21.00 | 21 | 20 | 1 | 0 | 0 |
| ..................................................GTAAGTGCACCAGATTGGGATT.................................................................................................... | 22 | 0 | 1 | 21.00 | 21 | 3 | 18 | 0 | 0 |
| ...................................................................................................TCCCAGTAATGTCCACTTACA.................................................... | 21 | 0 | 1 | 10.00 | 10 | 9 | 1 | 0 | 0 |
| ..................................................................................................TTCCCAGTAATGTCCACTTACA.................................................... | 22 | 0 | 1 | 8.00 | 8 | 6 | 1 | 1 | 0 |
| ...................................................................................................TCCCAGTAATGTCCACTTACAA................................................... | 22 | 0 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 |
| ...................................................................................................TCCCAGTAATGTCCACTTACAAGT................................................. | 24 | 1 | 1 | 5.00 | 5 | 4 | 1 | 0 | 0 |
| ..................................................GTAAGTGCACCAGATTGGGAT..................................................................................................... | 21 | 0 | 1 | 4.00 | 4 | 0 | 4 | 0 | 0 |
| ....................................................................................................CCCAGTAATGTCCACTTACAA................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ..................................................................................................................CTTACAAGGAGCCCCTGGATGGCTGGGT.............................. | 28 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 |
| ..................................................GTAAGTGCACCAGATTGGGA...................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 |
| ...................................................................................................TCCCAGTAATGTCCACTTT...................................................... | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................TTCCCAGTAATGTCCACTTACAA................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................TTCCCAGTGATGTCCACTTACT.................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................CCCAGTAATGTCCACTTACA.................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................TCCCAGTAATATCCACTTACAA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................................TCCCAGTAGTGTCCACTTACAAG.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................CCCAGTAATGTCCACTTACAAG.................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................TTCCCAGTAATGTCCACTTAC..................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................CCAGTAATGTCCACTTACAAGT................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................TCCCAGTAATGTCCACTTAC..................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................TAAGTGCACCAGATTGGGAT..................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................GTAACTGCACCAGATTGGGATT.................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................TGGCTGGGTGGACACCCGG.................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ................................................................TTTGGATCAACCTTTAAAAG........................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
GGACGCCCTCCGCCCGCTGGACGGTGACACGTAGAAGGCGGGCCGCTAGTCATTCACGTGGTCTAACCCTAATTGGAAGTTTTCGACGTCGAATTGATAAGGGTCATTACAGGTGAATGTTCCTCGGGGACCTACCGACCCACCTGTTGGATAAGCCTGGTAACCGGGAGAC
**************************************************(((((((.((...(((((((((((.((.((.....)).)).))))....)))))))...)).)))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528802 follicle cells |
M043 female body |
|---|---|---|---|---|---|---|---|
| .......................................................CCGTGGTCTAACCCTA..................................................................................................... | 16 | 1 | 4 | 0.25 | 1 | 1 | 0 |
| .........................................................................................................................................................AGCTTGGATACCGGGAGAC | 19 | 3 | 19 | 0.05 | 1 | 0 | 1 |