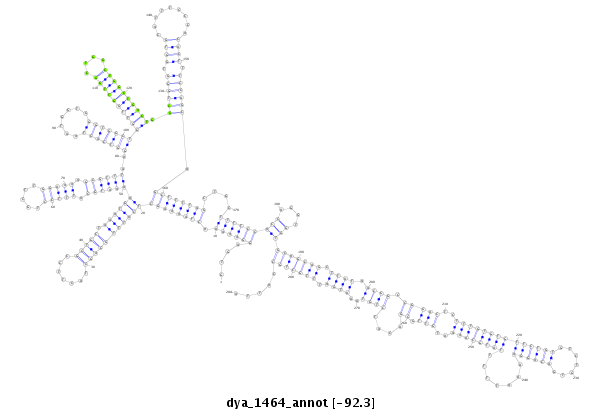

ID:dya_1464 |
Coordinate:X:17799846-17800133 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -92.2 | -92.0 | -91.8 |
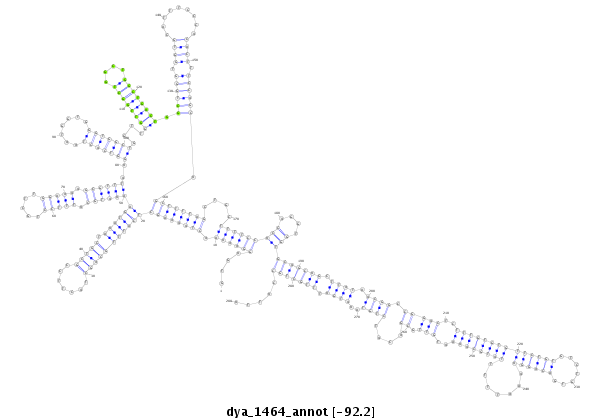 |
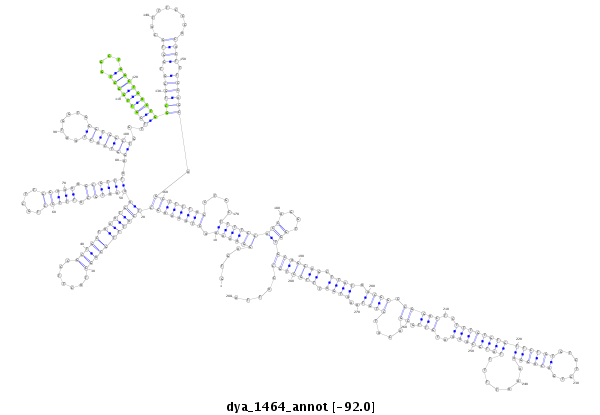 |
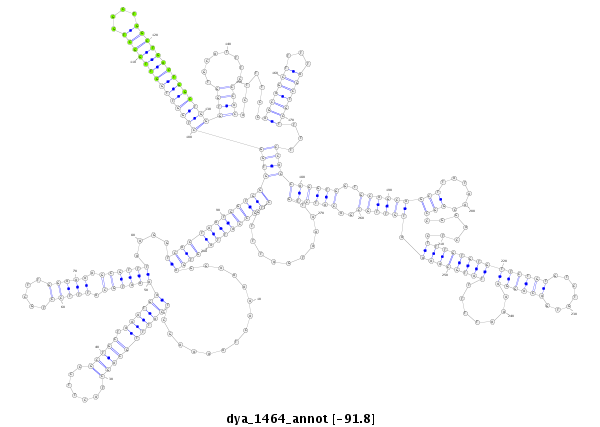 |
exon [dyak_GLEANR_16968:2]; exon [dyak_GLEANR_16968:1]; CDS [Dyak\GE15454-cds]; CDS [Dyak\GE15454-cds]; intron [Dyak\GE15454-in]
| Name | Class | Family | Strand |
| (CAGC)n | Simple_repeat | Simple_repeat | + |
| mature | star |
| ##################################################------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGAATCAGATGGACGATGGACCGGAAATGCGGACGCACGAGAACGCAAAGGTGAGGAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGGAAAAGCGTTTAAGGTCACTAATGGTGGGTGGCTGGTTGGTTGGCTGGCTGGCTGACTGGCTGGGTGGTGCATTTCCCACACTTTCCAGCAGCTTTTAGCTGCTTTTCCACCAGGTGGTGCAGCAGCTTATAACGGCCGGACTCTTTGTGTGTTCTGTGTCTGTGACAGGAAAATTTTTATGCAAAATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGATGGAGCGCATGGAGATTCCACGCTGGATGCCCTGGTCAGTGGCGC **************************************************....((((((.(((((((((((((((((((.......))))).))))))((((((.(((((.....))))).))))))..(((((((.......)))))))((((((((((....)))))))))).((((((.((((.........)))).)))))).))))))))....))))))(((....)))(((((((.((((.((((.(((((..((((((((((((((.......)))))).......))))))))..))).))....))))..)))).))))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V058 head |
V052 head |
M026 head |
M043 female body |
M056 embryo |
V046 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................................................................................................................................TCCACGCGGGGTGCCCTGGT......... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TGGTTGGTTAGCTAGCTGGATGA....................................................................................................................................................................................................................... | 23 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................TTTTGCGACATCTGTAATTGA............................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................GAAATGCGGACGCACAAG........................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................TGGCAGGGTGGTTGGCTGGCTG............................................................................................................................................................................................................................ | 22 | 2 | 14 | 0.43 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCAGGGGGGTTGGCTGGCT............................................................................................................................................................................................................................. | 21 | 3 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................GTTGGCTGGCTGGCTGACTGGC.................................................................................................................................................................................................................. | 22 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................TGGCGGGGTGGTTAGCTGGCTG............................................................................................................................................................................................................................ | 22 | 3 | 18 | 0.22 | 4 | 3 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCTGGGTGGTTAGCTGGCT............................................................................................................................................................................................................................. | 21 | 2 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................TGGCTGAGTGGCTAGCTGGTGC........................................................................................................................................................................................................ | 22 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................TGGCAGGGTGACTGGCTGGGTG............................................................................................................................................................................................................ | 22 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TGGTTGGTTGGCTAGCTGGCT......................................................................................................................................................................................................................... | 21 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................ACGGGTAATGCGGACGC................................................................................................................................................................................................................................................................................................................................................................ | 17 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TGGTTGGTTGTCGGGCTGGCTG........................................................................................................................................................................................................................ | 22 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........TGGACGGTGGACTGGTAATG....................................................................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 12 | 0.17 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCAGGGTGGTTGGCTGGCT............................................................................................................................................................................................................................. | 21 | 2 | 19 | 0.16 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................AAGCCTTTAAGGTAACTA.......................................................................................................................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................TGGCAGACTGGCTAGATGGTG......................................................................................................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TCGTTGGTTAGCTAGCTGGCTG........................................................................................................................................................................................................................ | 22 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TGGTTGGGTGGCTAGCTGGATGA....................................................................................................................................................................................................................... | 23 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................TTCCACCATGTGGTTCAGCT.................................................................................................................................................. | 20 | 3 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TCGTTGGTTGTCGGGCTGGCT......................................................................................................................................................................................................................... | 21 | 3 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................CTCATTGTTTGTTCTGTG................................................................................................................ | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................TGGCTGGGTGGTTGGCTGGCT............................................................................................................................................................................................................................. | 21 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCGGGGTGGTTGGCCGGCTG............................................................................................................................................................................................................................ | 22 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGCCTGGGTGGTTCGCTGGCTG............................................................................................................................................................................................................................ | 22 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCGGGGTGGTTGGCTGGCT............................................................................................................................................................................................................................. | 21 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCTGGGTGGTTAGCTGGCTG............................................................................................................................................................................................................................ | 22 | 2 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................AGGGAGGATGGTTGGTTGGCT................................................................................................................................................................................................................................. | 21 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCGGGGTGGTTCGCTGGCTG............................................................................................................................................................................................................................ | 22 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................TGGCCGGGGGGCTGACTGGCT................................................................................................................................................................................................................. | 21 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCAGGGAGGTTGGCTGGCTG............................................................................................................................................................................................................................ | 22 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCCGGGTGGTTGGCTGGCT............................................................................................................................................................................................................................. | 21 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TGGTTGGTTGGCTAGCTGGGT......................................................................................................................................................................................................................... | 21 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCAGGGTGGTTTGCTGGCTG............................................................................................................................................................................................................................ | 22 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCTGTGTGGTTGGCTGGCTTG........................................................................................................................................................................................................................... | 23 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................AGGCGGGGTGGTTGGCTGGCT............................................................................................................................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................TGGCAGGGTGGCGGACTGGCT................................................................................................................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCGGGGTGGTTAGCTGGC.............................................................................................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCTGGGTGGTTGGCTGGGTGG........................................................................................................................................................................................................................... | 23 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCCGGGGGGTTGGCTGGCT............................................................................................................................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCTGCGTGGTTGGCGGGCTG............................................................................................................................................................................................................................ | 22 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCCGGGTGGTTGGCTGGCCG............................................................................................................................................................................................................................ | 22 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................TGGCCGGTGGACTGGCTGGG.............................................................................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TGGCTGGGTGGTTAGCAGGCT............................................................................................................................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
ACTTAGTCTACCTGCTACCTGGCCTTTACGCCTGCGTGCTCTTGCGTTTCCACTCCTTTTTGATTTTCGAGTAAACGTCGATGAAGGCGACGATTTACTTTTACGTAAAGGACGAACCTTTTCGCAAATTCCAGTGATTACCACCCACCGACCAACCAACCGACCGACCGACTGACCGACCCACCACGTAAAGGGTGTGAAAGGTCGTCGAAAATCGACGAAAAGGTGGTCCACCACGTCGTCGAATATTGCCGGCCTGAGAAACACACAAGACACAGACACTGTCCTTTTAAAAATACGTTTTACAACGCTGTAGACATTTACTAAACGACGGTAATCTTCTACCTCGCGTACCTCTAAGGTGCGACCTACGGGACCAGTCACCGCG
**************************************************....((((((.(((((((((((((((((((.......))))).))))))((((((.(((((.....))))).))))))..(((((((.......)))))))((((((((((....)))))))))).((((((.((((.........)))).)))))).))))))))....))))))(((....)))(((((((.((((.((((.(((((..((((((((((((((.......)))))).......))))))))..))).))....))))..)))).))))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V058 head |
V046 embryo |
|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................................................................................GACACTGTCCTATTAAA.............................................................................................. | 17 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| ....................................................................................................................................................................................CCACCACGGAAAGGATGTGT............................................................................................................................................................................................ | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 |
| .................................................................................................................................................................................................................................................................................CGCCGACACTGTCCTGTTAAA.............................................................................................. | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 |
| ..................................................................................................................................................................................................................................................................................AGAGACACTGTCCTGTTATA.............................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 1 |
| .....................................................................................................................................................................................................................................................................................GACACTGTCCTTTGATA.............................................................................................. | 17 | 2 | 14 | 0.07 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droYak3 | X:17799796-17800183 - | dya_1464 | TG-----------------AA-----------------------TCAGATGGAC---GATGGACCGGAAATGCGGAC-GCACGAGA---AC------------GCAAAGGTGAGGAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGGT--CAC----------T-------------------------------------------AAT----------------------------------GGTGGGTGGCTGGTTGGTTGGC------------TGGCTGGCTGACTGGCTGGGTGGTGC-------------------------------------------------------------------------------A---------TTT-CCCA-----------C--ACTTTCCAGCAGC--------------TTTT-AG-CTGCTTTTCCACCAG-------------------G------TGGTGC------------------A---------GCA-----GCTTATAACGGCC------------------------------------------------------GGACT-CTT-----------------------------------TGTGTGTTCTGTGTC-TGT--GACAGGAAAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAT------------------------------------------------GG----------------------AG------CGCATGGAGATTC-----------------------CACG--CTGG----ATGCC-----------CTGGTCAGTGGCG-------C-------- |
| droEre2 | scaffold_4690:5912827-5913175 + | CG-----------------AA-----------------------TCAGGTGGAC---GGTGGACCGGAAATGCGGAC-GCAGGAGA---ATGCCACCAG--GAGCAAAGGTGAGGAAAAACTAAAAGCTCATTTGCAGCTACATCCGCTGCTAAATGAAAATGCATTTCCTGCCTGG--AA-AAGCGTTTAAGGT--CAC----------T-------------------------------------------AAT----------------------------------GGTGGGTGGC----------------------------------------TGTGTGGTGC-------------------------------------------------------------------------------CCACCCCCACTT-----------TTC---CAGCTTTTC--------------------------------------CAGCAG-------------------G------TGGTGC------------------A---------GCA-----GCTTCTAACGGTC------------------------------------------------------GGACT-CTTG----------------------------------------TG--TTGTC-TGT--GACAGC-AAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAT------------------------------------------------GG----------------------AG------CGCATGGAGATTC-----------------------CACG--CTGG----ATGCC-----------CTGGTCAGTGGTG-------C-------- | |
| droSec2 | scaffold_68:97766-98115 - | TT-----------------GA-----------------------CCAGGTGGAC---GGTGGACCGGAAATGCGGAC-GCACGACA---ACGCCGCCAG--GAGCGAAGGTGAGGAAAAACTAAAAGCTCATTCGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGGT--CAC----------T-------------------------------------------AAT----------------------------------GGTGGGTGGC----------------------------------------TGGGTGGTGC--------------------------------------------------------------------------------CACCCCCGCTT-----------TTC---CAGCTTTTC--------------------------------------CATCAG-------------------G------TGGTGC------------------A---------GCA-----GCCTCTAACGGCC------------------------------------------------------GGACC-CTTG----------------------------------------TG--TTGTC-TGT--GACAGC-AAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAT------------------------------------------------GG----------------------AG------CGCATGGAGATTC-----------------------CGCGCCCTAG----ATGCC-----------CTGGTCAGTGGTG-------C-------- | |
| droSim2 | x:8753401-8753750 - | dsi_28975 | TT-----------------GA-----------------------CCAGGTGGAC---GGTGGACCGGAAATGCGGAC-GCACGACA---ACGCCGCCAG--GAGCAAAGGTGAGGAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGGT--CAC----------T-------------------------------------------AAT----------------------------------GGTGGGTGGC----------------------------------------TGGGTGGTGC--------------------------------------------------------------------------------CACCCCCGCTT-----------TTC---CAGCTTTTC--------------------------------------CACCAG-------------------G------TGGTGC------------------A---------GCA-----GCCTCTAACGGCC------------------------------------------------------GGACT-CTTG----------------------------------------TG--TTGTC-TGT--GACAGC-AAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAT------------------------------------------------GG----------------------AG------CGCATGGAGATTC-----------------------CACGCCCTGG----ATGCC-----------CTGGTCAGTGGTG-------C-------- |
| dm3 | chrX:9206559-9206913 - | TG-----------------AA-----------------------CCAGGTGGAC---GGTGGACCGGAAATGCGGAC-GCACGAGA---ATGC-----------CGAAGGTGAGGAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGGT--CAC----------T-------------------------------------------AAT----------------------------------GGTGGGTGGC----------------------------------------TGGGTGGTGC--------------------------------------------------------------------------------CACCACCGCTT-----------TTC---CACATTTTCCAGGAA--------------------------------AGCCAG-------------------G------TGGTGC------------------A---------GCA-----GCATCTAACGGCC------------------------------------------------------GGACT-CTTG----------------------------------------TG--TTGTC-TGT--GACAGC-AAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAT------------------------------------------------GG----------------------AG------CGGATGGAGATTC-----------------------CACGCCCTGG----ATACC-----------CTGGTCGATGGTG-------CACCAGTGG | |
| droEug1 | scf7180000409032:750676-751047 + | AT-----------------GA-----------------------TCAGGTGGAC---GATGGACCGGAAATGCGGAT-GCACGAGATGCACGCCGCCAG--GAGCGAAGGTGAGCAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGGT--CAA----------T-------------------------------------------AAC----------------------------------GGTGTATGGT------------------------------------T---TGGGTGGTGC-------------------------------------------------------------------------------A--------CTT-----------TCC---CCGATTTTCC-------------------------------------AACCAG-------------------G------TGGTGC---TACT-----------A---------GCA-----GCGTCTAACGGTC------------------------------------------------------GG-CT-CTTC----------------------------------------TG--TTGTC-TGT--TTCAGG-AAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAA------------------------------------------------GG----------------------AG------CGCATGGAGCTGCTGGAGCTGTTGGAGCCTACATCCT--GCACTGG----ATCCT-----------ATGGTCTGTGGTG-------C-------- | |
| droBia1 | scf7180000301760:5177271-5177589 - | AG-----------------GG-----------------------CCAGGTGGCC-------GACCGGAAATGCGGAT-GCACCAGA---GCGCCGCCAG--GAGCGAAGGTGAGCAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCCAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAG-T--CAA----------T-------------------------------------------AAT----------------------------------GCTGGCTGGC------------------------------------TGGATGGGAGGTGC-------------------------------------------------------------------------------A--------CTC-----------GTC---CA-CTTTCC--------------------------------------CACCAG-------------------G------TGGTGC------------------A---------GC------------------C------------------------------------------------------AGACT-CC-C----------------------------------------TG--TTGTC-TGT--GACGGC-AAATTTTT-ATGCTA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAT------------------------------------------------GG----------------------AG------CGCATGGAGATTCGGGA---------GCCT--------------GG------CCCCAGG-----------CCA-------------T-------- | |
| droTak1 | scf7180000415696:169747-170051 - | AG-----------------GATCT---------------------GA----------GGTGGACCGGAAATGCGGAT-GCACGAGA---ACGCCGCCAGGTGAGCGAAGGTGAGCAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGCTACCAA----------T-------------------------------------------AAT----------------------------------GGTGGGTGGT-----------------------------------------------------------------------------------------------------------------------CTAGG--------------CTTTCCCCGCAGTTTTCCCCCAG-CTTTC--------------------------------------CA---------------------------------------------------------------------------------------------------------------------------------------------ACT-CTCG----------------------------------------TG--TTCTC-TGT--GACAGC-AAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAT------------------------------------------------GG----------------------AGATG-------TGGAGGAGCA------------------------------GG----TTG-C-----------CTGGTTA-----G-------C-------- | |
| droEle1 | scf7180000491018:50698-50969 + | AG------------CTGGTAA-----------------------ACAGGTGGACCTGGTTGCACCGGAAATGCGGAT-GCACGAG-----------------ATCGAAGGTGAGCA-AAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGT--GA-AAGCGTTTAAGGT--CAG----------TC--------------------------------------ACTGAAT----------------------------------GGTTG---G-------------------------------------------------------------------------------------------------------------------------------------------CTTT-T--------CCC---CA-CTTTTC--------------------------------------CAGCAGACAGCAGATAGGAGTTAGG-AGATGG--------GA------------------------------------------------------------------------------------------------------------------------------------------------------------GA-TGG--G------AGATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAA------------------------------------------------GG----------------------AG------C---------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000780067:101495-101810 - | AG-----------------CATCAG---------GAGC-AGGA-ACAGGTGGAC---GTTGCACCGGAAATGC-----------------------CAG--GAGCGAAGGTGAGCAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGGT--CAG--------------------------------------------------------T----------------------------------GGTGGGTGGC--------TGGA------------CAAA--------TGGATGGGTGGTGC-------------------------------------------------------------------------------ACTCTGCCACTT-----------TCC---CA-CTTTTC--------------------------------------CACCAG-------------------G------TGGTGGTCC-ACTC---------------------------------------------------------------------------------------------------TAGTGC----------------------------------------CG--TTGTC-TGT--GACAGC-AGATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAA------------------------------------------------GG----------------------AGTTGTGCCCCATGGATGTTC---------------------------------------------------------------------------------- | |
| droFic1 | scf7180000454073:2119309-2119696 - | TG-----------------GA-----------------------C--GGTGGAC---GGTGGACCGGAAATGCGGAT-GCACCAGG---ACGAAGCCAG--GAGCGAAGGTGAGCAAAAACTAAAAGCTCATTTGCAGCTATTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGGC--CAT----------TG--------------------------------------AATGAAT----------------------------------GG---CTGCGTGGTTGGGTGGCTCCATGGTTGGATTGG--------TGGTTAGGTGGTGC--------------------------------------------------------------------------------------------------------C---CC-TTTTTG--------------------------------------CAGCAG-------------------G------T---GC---AACT-----------T-----------T-----GCGTCTAACGGTC-------------------------------------------------------GACT-CCTC----------------------------------------TG--TTGTC-TGTGTGACGGC-AAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAAG---------------------------------------------CTGG----------------------AG---------ATGGAGCACCA--AGCTCCAGGAGCCT-------ACGTTGTGG----ATCCT-----------CTGGCCAATTG---------GACCCTCGG | |
| droKik1 | scf7180000302592:15860-16234 + | C--------------------------------------------------------GGTTAACCGGAAATGCGTACTGCAAAAGA---GCGCCGTCAG--GAGCAAAGGTGAGCAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGGT--CAGG--CCGTCGTGATGGCGCGGTGCGGGGCTGGGGGGGTTGTGGACCCCGAGGACCAGT----------------------------------GGT-----------TGCGTGGC------------CAACTGGCTGAGTGGGCGAGCGGTGC-------------------------------------------------------------------------------A-----------------------------------------------------------------------------------------------------------------------------------------------------TCGTCTAGCGGTC----------------------------------------------------------TCCTCC----------------------------------------TG--CTGTC-TGT--GACAGC-AAATTTTT-ATCCAA-AGCGTTGCGACATCTGTAAATGATTTGCTCCCATTAGAAGATGGGCC----------------CCCAAAAAGGAGGTGGGAAGGCAACCAGG----------------------AG------TTTATGGAGATTCAGGA---------GCCT----------------------------------------------------------------- | |
| droAna3 | scaffold_13334:358800-359176 + | AT-----------------AA------------------------CAGGATAAC---GGCGGACCGGAAATGC------------------GCCGCCAG--GAGCAAAGGTGAGCAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGGCTGG--GAAAAGCGTTTAAGGT--CAG----CAGCATT-------------------------------------------AATGCCGAGAACTCACCCTGTTCCAGGTCCTGGTCCTGGTC-------------------------------------------------------------------CTG----ATC----------------------------------------------------------------------------------------------------------CTGTAAATCCTTTTT-AGTC----------------------TAGTG---ATGGCTATGGTGGTGATGTAAT-----------CGG-------------------AATAACCACCTCCTCCTCCA----------------------------------------------ACTCCTCC---------------------------AGTGGCCAGTG--CC--TTGTC-TGT--GACAGGAACATTTTT-ATGCAA-AATGCTGCGACATCTGTAAATGATATGCTGCCATTAGGAGGA------------------------------------------------GA--------------------------------------------------------GCCTACAAC----GCACTCGAAGGACACC-----------T------------------TC-------- | |
| droBip1 | scf7180000396431:1195065-1195424 - | TG-----------------ACTGA---------------------GAGGACAAC---GGCGGACCGGAAATGC------------------GCCGCCAG--GAGCAAAGGTGAGCAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGGCTGGGAAA-AAGCGTTTAAGGT--CAGAAAACAGCATT-------------------------------------------AATGCCGAGTACTCACCCTGATC----TCCT------GGTGGAT-------------------------------------------------------------------------C----------------------------------------------------------------------------------------------------------CTGCAAATCCATTTT-AGGCTGCT----------------GCTAGTGGTAATG-GTATGG--------CAAC-----------CGG-------------------AATAACGACCTCCTCC--------------------------------------------------ACT-C-------------------------------------GGTGCGTT--TTGTC-TCT--GACAGGAAAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATATGCTGCCATTAGGAGGA------------------------------------------------GA--------------------------------------------------------GCCTACAAC----GCACTCGAAGGACACC-----------T------------------TC-------- | |
| dp5 | XL_group1a:7801378-7801665 + | G------------------AGCCAG---------GAGCCAGGAGCCA-------------------GGG-------------------------ATCCG--GAGTGAAGGTGAGCAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGC---CAT----------TG--------------------------------------C------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACC-------------------------------------------------------------------------------------------------------------------------------GCCATCGCCAT-------------------------------------CGCCATGGTAGTGGGAGGTTTCACCGCTCCTCGTGCCAGCCTCCTCC----------------------------------------TG--TTGTC-TGT--GCTAG--AAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGCA------------------------------------------------GG--------------------------------------------------------G-------------CCCTAG------------GCCTATGCCTGGTCCATGGTT-------A-------- | |
| droPer2 | scaffold_15:151661-152000 + | TTGCACAATTTGTGCTAGTAGCCAGGGATCCAGGGAGCCAGGAGCCA-------------------GGG-------------------------ATCCG--GAGTGAAGGTGAGCAAAAACTAAAAGCTCATTTGCAGCTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTAAGC---CAT----------TG--------------------------------------C-----C--------------------------------------------------------------------------------------------A-----------------------------CCGCCACCGCCATCAC-------------------------------------------------------------------------------------------------------------------------------------------------CATC------------------A---------CCA-----TC-----------------------ACCATCGCCATGGTAGTGGGAGGGTTCACCGCTCCTCTTGCCAGCCTCCTCC----------------------------------------TG--TTGTC-TGT--GATAG--AAATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGCA------------------------------------------------GG--------------------------------------------------------G-------------CCCTAG------------GCCTATGCCTGGTCCATGGTTAGGCCATC-------- | |
| droWil2 | scf2_1100000004909:2870354-2870695 + | --------------------------------------------------------------ACCGGAAATGCAAATTGCA---------CGCAGTCAAG-AAGAGAAAGTGAGGAAAAACTAAAAGCTCATTTGCAGTTACTTCCGCTGCTAAATGAAAATGCATTTCCTGCTTGG--AA-AAGCGTTTGAGTT---TT----------T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGAATAA-CCTACCAACATATGTATGCAAATGTTTGTTGTGTG----------------------GAGTG---AAGG------TGGATC------------------A---------GGA-----GC-----------------------AGGATCGGGA-------------------------------------------------------------------------GTAAAGG--AG--TTGTC-TGT--GAC------ATTTTTAATGCAAAAATGTTGCGACATCTGTAAATGATTTGCTACCATTAGAAAATACGCCAGGCAGTTGCGTATCGTCCGAGATGGAGTTGGAG-----------ATGGAGTCGGAGATGAAGTCGGAG---------A----------------------------------------------------------------------TGGAG-------C-------- | |
| droVir3 | scaffold_12970:8580857-8581150 + | -----------------------------------------------------------------------------------------------------------AAGTAGGCGAAAACTAAAAGCTCATTTGCAGTTACTTCCGCTGCTAAATGAAAATGCATTTCCTGGCTGG--AAGAGGCGTTTAAGTT---G----CCGCCATTATGGTACG------------------------------------------A-------------------------------------------------------------------------------------GTGCCACCCCTCTGTCTACTCCATCCTTGCACCCGCCGCCGCCGCCGCCGCCGCCGCCACCGAAGTAAATGTTCCAATGTTGCC---------------------------------------------------------------------------------------------------------------------------------------------T-C-----GCATGTAAC-------------------------------------------------------------------------------------------------------------G--GTGTC-TAT--GAC------ATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTGCCATTAGAAGAAA---------------------------------------------CAGGCCCGAATCAGAGT----------------------TAAAATTT-----------------------G-----------------------------------------A-------C-------- | |
| droMoj3 | scaffold_6482:676558-676827 - | -------------------------------------------------------------------------------------------------------------------AAAAACTAAAAGCTCATTTGCAGTTACTTCCGCTGCTAAATGAAAATGCATTTCCTGGCTGG--AAGAGGCGTTTAAGTT---G----CCGCCATTATGGAAAG-----------GGATGGAAGTGGACCGCC-GTAC-------------------------------------------------------------------------------------------------------------------CATTGGAGCACCAG------------------------------------------------------------------------------------------------------------------------------------------------------------------CCG-ACTCCATTGGCATTGGCATTGGCATTGGGATTCCA------------------------------------------------------------------------TG-----------------------------------TGA--CG--CTGTCGTAT--GAC------ATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGGTGCCATTAGAAGAAGAATA-----------------------------------------CATGCTCGGATCTGAGT-------------------------------------------------------------------------------------------TAGAA-------C-------- | |
| droGri2 | scaffold_15203:11339509-11339798 + | ----------------------------------------------------------------------------------------------------------AAAGCGAACCAAAACTAAAAGCTAATTTGCAGTTACTTCCGCTGCTAAATGAAAATGCATTTCCTGGCTGA--AAGAGGCGTTTAAGT---CG----CCGCCATTATGGAACG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GG-------------------TATACCAATCTCCTTCTCCCTCTCCCTCTCC--------------------CTCTCCT---------CT-CTCTCTCTCTCTCTAAGGCTGCTACGGATGCA--CGCCAAAA--CG--GTGTC-TGT--GAC------ATTTTT-ATGCAA-AATGTTGCGACATCTGTAAATGATTTGCTACCATTAGAAGAAG---------------------------------------------CTGGCTCGAATCAGAGT----------------------T-ACATTC-------------------------------------------------------GGACACCGGCT-------T-------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 12:59 AM