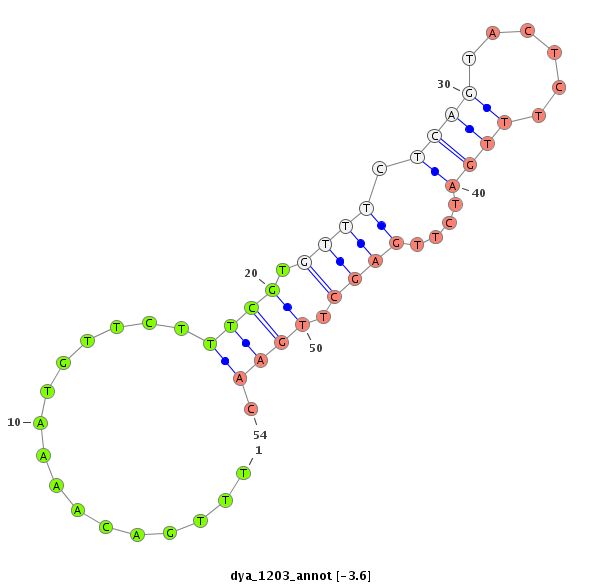
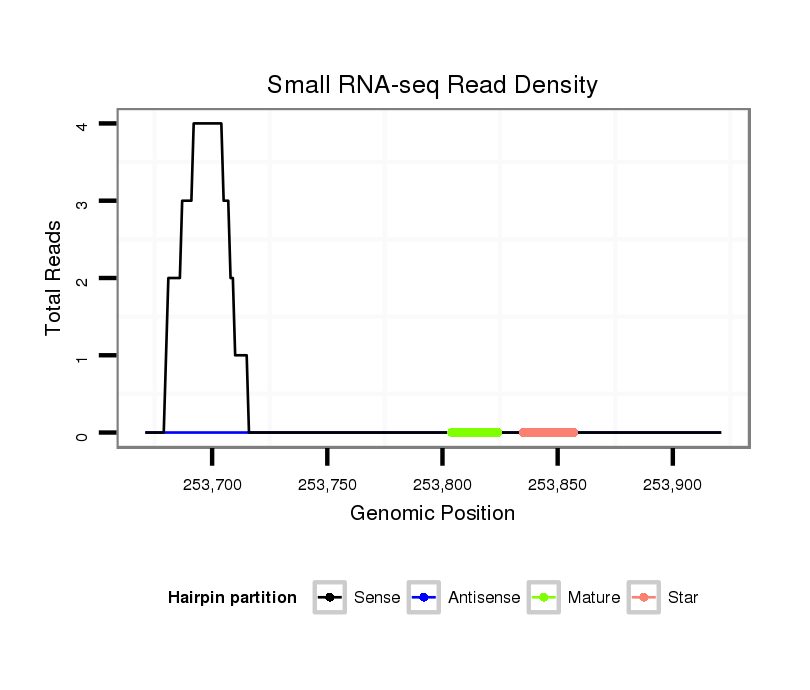
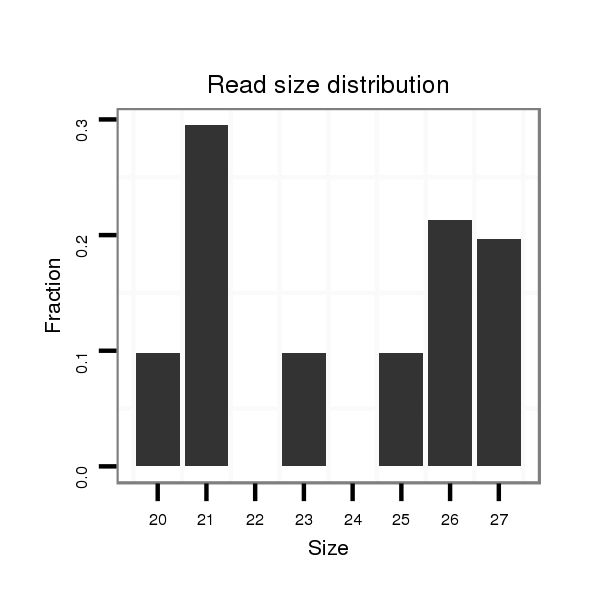
| AAAAAAG-------------GG---------CACGTAGGGCATTAGTCAAACTACATTGTACACTAGTC------------------------------------------------------------------------------GGCATATATATTTTGACATAG--CGATGCTTCATTTTCTTTTTCAAT---TATTTCTTTATTTAAAGGAAAACAATTAGTGGTTTAATTTTTAATAGATATTAGTAATG-----------------------------------------------------------------------------------------------TATTACT----------------- | Size | Hit Count | Total Norm | Total | M044
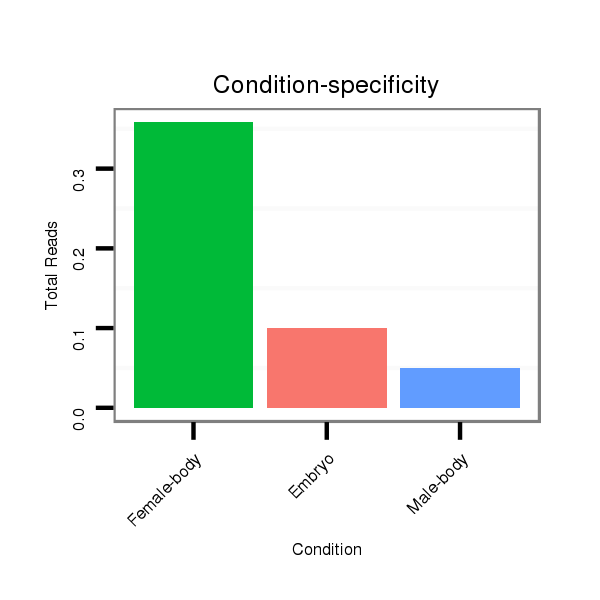
Female-body | M058
Embryo | V039
Embryo | V055
Head | V105
Male-body | V106
Head |
| ....................................................................................................................................................................................................................AGGAAAACAATTAGTGGTTTAATTT.......................................................................................................................................... | 25 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................ATATTTTGACATAG--CGATGCT...................................................................................................................................................................................................... | 21 | 20 | 0.35 | 7 | 2 | 0 | 0 | 0 | 5 | 0 |
| ...............................................................CTAGTC------------------------------------------------------------------------------GGCATATATATTTTG..................................................................................................................................................................................................................... | 21 | 20 | 0.35 | 7 | 2 | 0 | 0 | 0 | 0 | 5 |
| ..............................................................................................................................................................................................................................................TAATAGATATTAGTAATG-----------------------------------------------------------------------------------------------TATTACT----------------- | 25 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TATATTTTGACATAG--CGATGC....................................................................................................................................................................................................... | 21 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 4 | 0 |
| ..............................................................................................................................................................TTTGACATAG--CGATGCTTCAT.................................................................................................................................................................................................. | 21 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................TTTTGACATAG--CGATGCTTCA................................................................................................................................................................................................... | 21 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................ATTTTGACATAG--CGATGCTTC.................................................................................................................................................................................................... | 21 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 3 | 0 |
| ........................................................................................................................................................ATATATTTTGACATAG--CGATG........................................................................................................................................................................................................ | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| ..................................................................................................................................................................ACATAG--CGATGCTTCATTTTC.............................................................................................................................................................................................. | 21 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................ATATATTTTGACATAG--CGATGC....................................................................................................................................................................................................... | 22 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 1 | 1 |
| ................................................................................................................................................................................................................................................ATAGATATTAGTAATG-----------------------------------------------------------------------------------------------TATTA................... | 21 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TATATTTTGACATAG--CGATGCTT..................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TATATTTTGACATAG--CGATGCTTCATT................................................................................................................................................................................................. | 27 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TACACTAGTC------------------------------------------------------------------------------GGCATATATATTTTGACA.................................................................................................................................................................................................................. | 28 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................ATATATTTTGACATAG--CGAT......................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................TATATATTTTGACATAG--CGA.......................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................CACTAGTC------------------------------------------------------------------------------GGCATATATATTTTGACA.................................................................................................................................................................................................................. | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................CATAG--CGATGCTTCATTTTCT............................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................GTC------------------------------------------------------------------------------GGCATATATATTTTGAC................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................TTGACATAG--CGATGCTTCAT.................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................TTCTTTTTCAAT---TATTTCTTT......................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................ATGCTTCATTTTCTTTTTCG....................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................CATATATATTTTGACATAG--CG........................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................TTTCTTTTTCAAT---TATTTCTT.......................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................TGACATAG--CGATGCTTCATTTTCTTC........................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................CTAGTC------------------------------------------------------------------------------GGCATATATATTTTGA.................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................................ATATATTTTGACATAG--CGATT........................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................ATATATTTTGACATAG--CGATC........................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................TACACTAGTC------------------------------------------------------------------------------GGCATATATA.......................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TATATATTTTGACATAG--CGAA......................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................TATATATTTTGACATAG--CGATG........................................................................................................................................................................................................ | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................................CTTCATTTTCTTTTTCAAT---T................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |