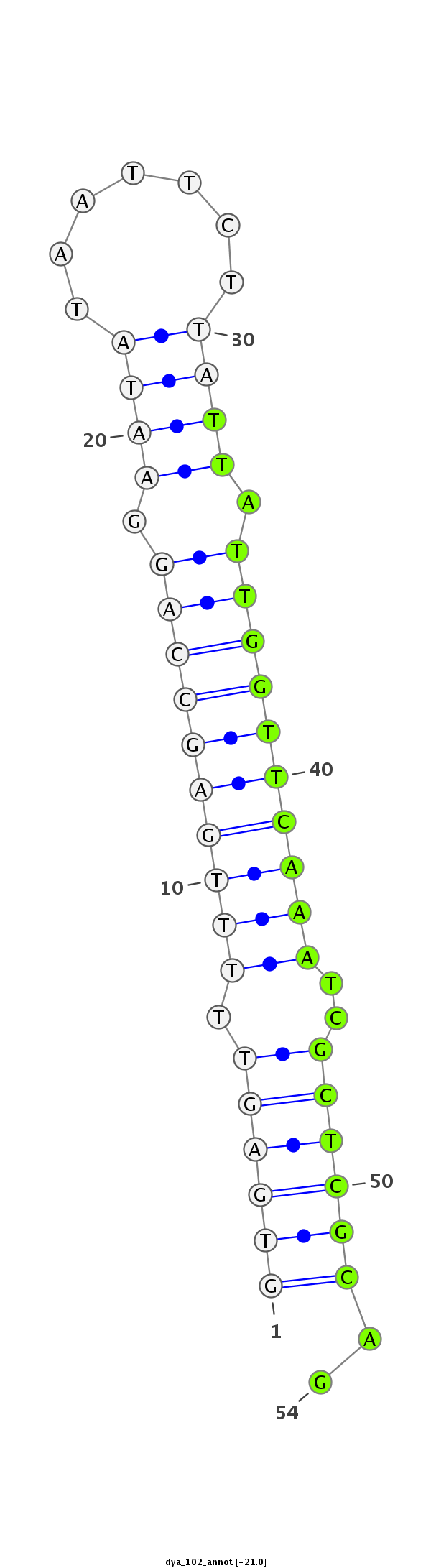
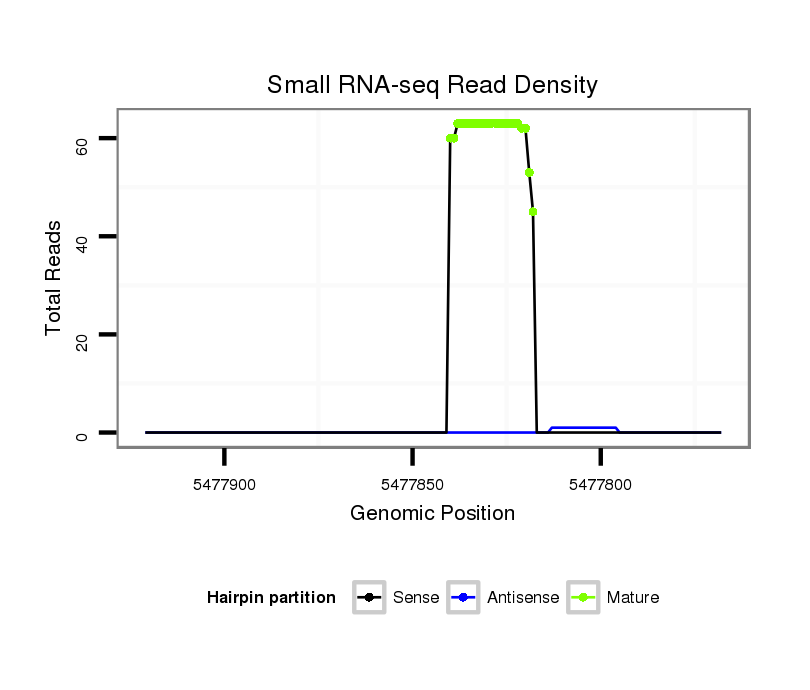
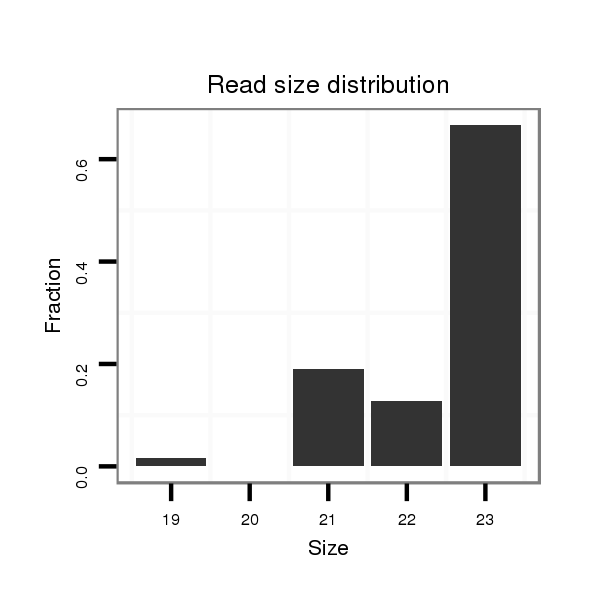
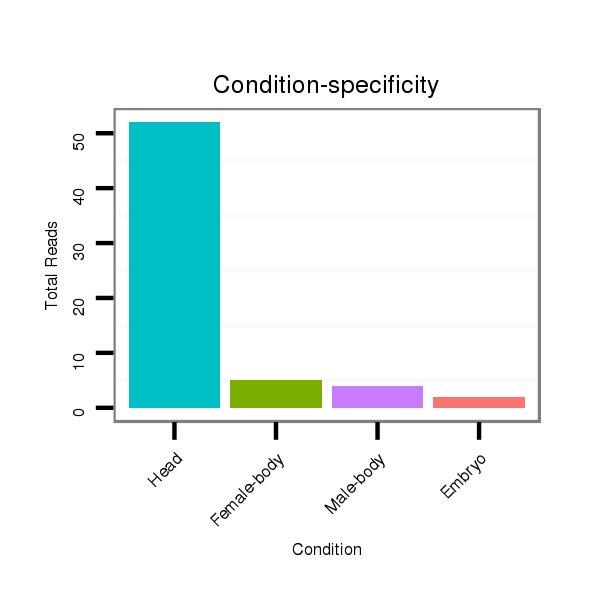
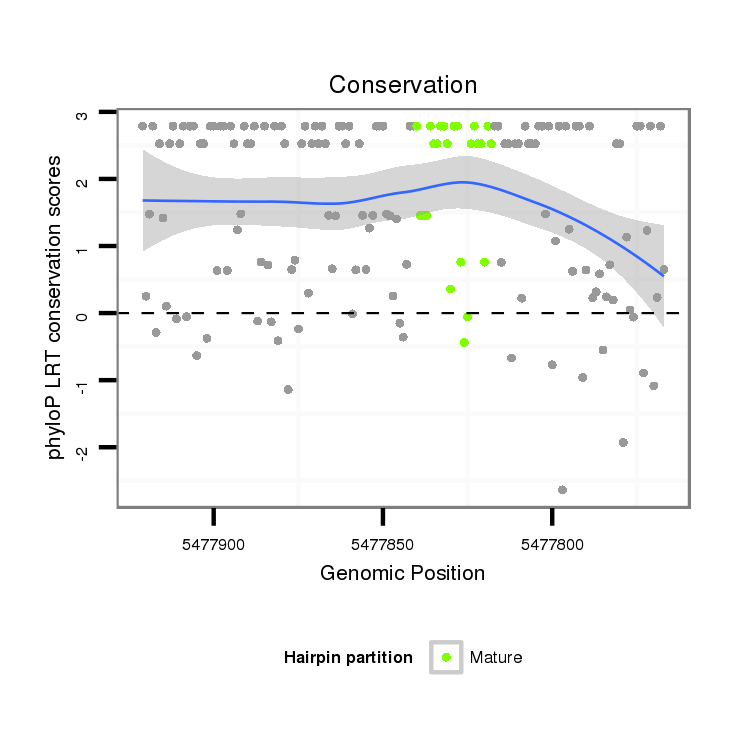
ID:dya_102 |
Coordinate:3R:5477818-5477871 - |
Confidence:Known Ortholog |
Class:Mirtron |
Genomic Locale:intron |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.6 | -20.2 |
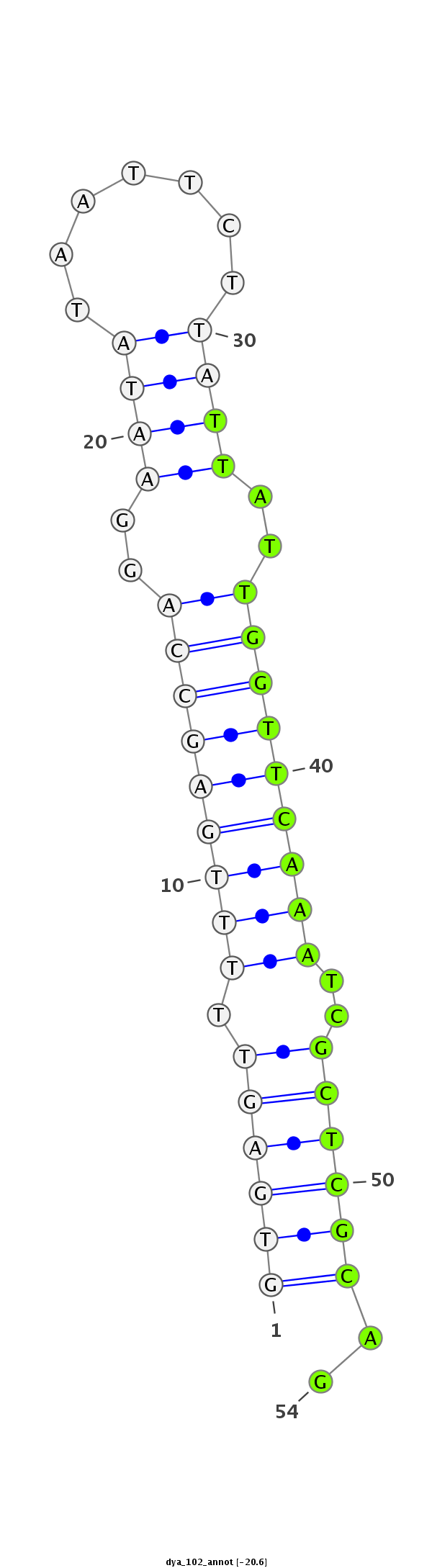 |
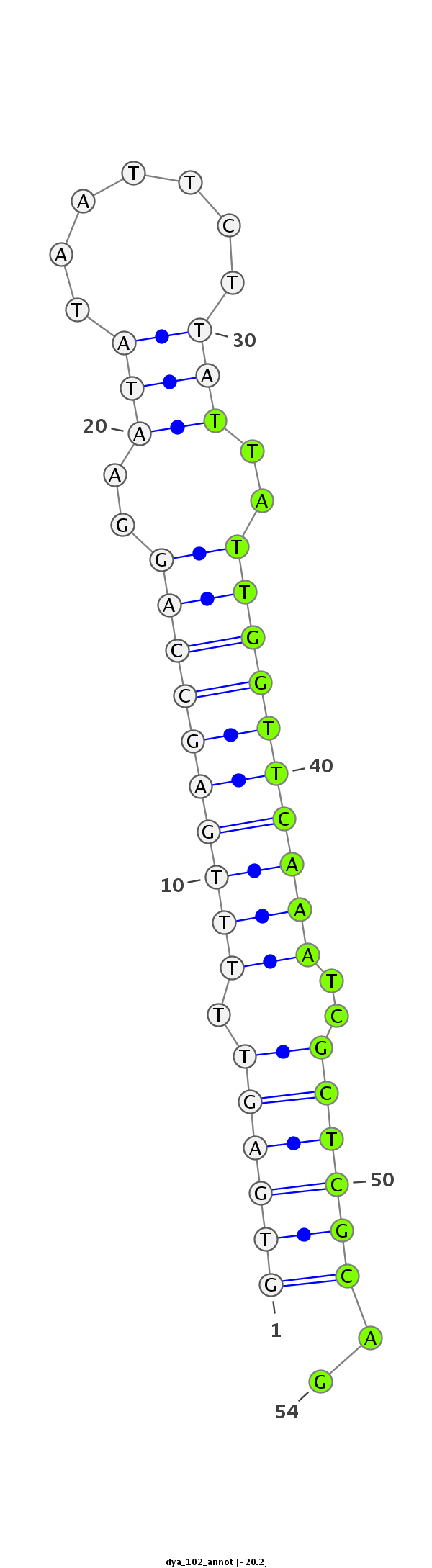 |
intron [Dyak\GE25033-in]; utr3 [utr3_minus_8261]; CDS [Dyak\GE25033-cds]; CDS [Dyak\GE25033-cds]
No Repeatable elements found
|
ACATCGAGCACCACTTTCGCAACATAACCATGGACATTCTGACCGTGCATGTGAGTTTTTGAGCCAGGAATATAATTCTTATTATTGGTTCAAATCGCTCGCAGAATCCACCCTGGCAAATCCTCACCAAAAACAGCCATGGAGTCATCGTTGA
**************************************************((((((.((((((((((.((((.......)))).))))))))))..))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M026 head |
V058 head |
M056 embryo |
V052 head |
M043 female body |
V120 male body |
V046 embryo |
GSM1528802 follicle cells |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................TTATTGGTTCAAATCGCTCGCAG.................................................. | 23 | 0 | 1 | 42.00 | 42 | 30 | 7 | 1 | 0 | 4 | 0 | 0 | 0 |
| .................................................................................TTATTGGTTCAAATCGCTCGC.................................................... | 21 | 0 | 1 | 9.00 | 9 | 7 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| .................................................................................TTATTGGTTCAAATCGCTCGCA................................................... | 22 | 0 | 1 | 8.00 | 8 | 1 | 3 | 0 | 2 | 1 | 1 | 0 | 0 |
| ...................................................................................ATTGGTTCAAATCGCTCGCAGT................................................. | 22 | 1 | 1 | 6.00 | 6 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 |
| .................................................................................TTATTGGTTCAAATCGCTCGCAGT................................................. | 24 | 1 | 1 | 4.00 | 4 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................ATTGGTTCAAATCGCTCGCAG.................................................. | 21 | 0 | 1 | 3.00 | 3 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................AGTGGTTCAAATCGCTCGCAGT................................................. | 22 | 2 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AGCCAGGTATATTATTCCTATT....................................................................... | 22 | 3 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................AGCCAGGTATATAATTCCTATG....................................................................... | 22 | 3 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................ATTGGTTCAAATCGCTCGCAGG................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TTATTGGTTCAAATCGCTAGCAG.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................ATTGGTTCAAATCGTTCGCAGT................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AGCCAGGTATATAATTCCTATC....................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TTATTGGTTCAAATCGCTC...................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................ATTGGTTCAAATCGCTCGCAGC................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TTATTGGTTCAAATCGCTCGCCGC................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TTATTGGTTCAAATCTCTCG..................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................CTATTGGTTCAAATCGCTCGCAG.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AGCCAGGTATATAATTCCTATT....................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................TTATTGGTTCAAATCGCTCGCCG.................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................AGCCAGGGATATAATTCCTATC....................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AGCCAGGGATATTATTCCTAT........................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TTATTGGGTCAAATCGTGCGC.................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TTATTGGTCCAAATCGCTCGCAG.................................................. | 23 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AGCCAGGGATATAGTTCCTA......................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................TAGTGCTTGAAATCGCTCGC.................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................AAATCGGTCGCAGAGTC.............................................. | 17 | 2 | 18 | 0.17 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
|
TGTAGCTCGTGGTGAAAGCGTTGTATTGGTACCTGTAAGACTGGCACGTACACTCAAAAACTCGGTCCTTATATTAAGAATAATAACCAAGTTTAGCGAGCGTCTTAGGTGGGACCGTTTAGGAGTGGTTTTTGTCGGTACCTCAGTAGCAACT
**************************************************((((((.((((((((((.((((.......)))).))))))))))..))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V058 head |
V046 embryo |
|---|---|---|---|---|---|---|---|
| ............................................................................................................GTGGGACCGTTTAGGAGT............................ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ....GCTCGTGCTGAGAGCGTTG................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| ................................CTGTAAGTCTGGCAGGAACA...................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 |