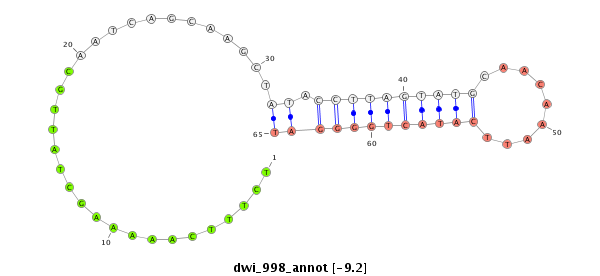
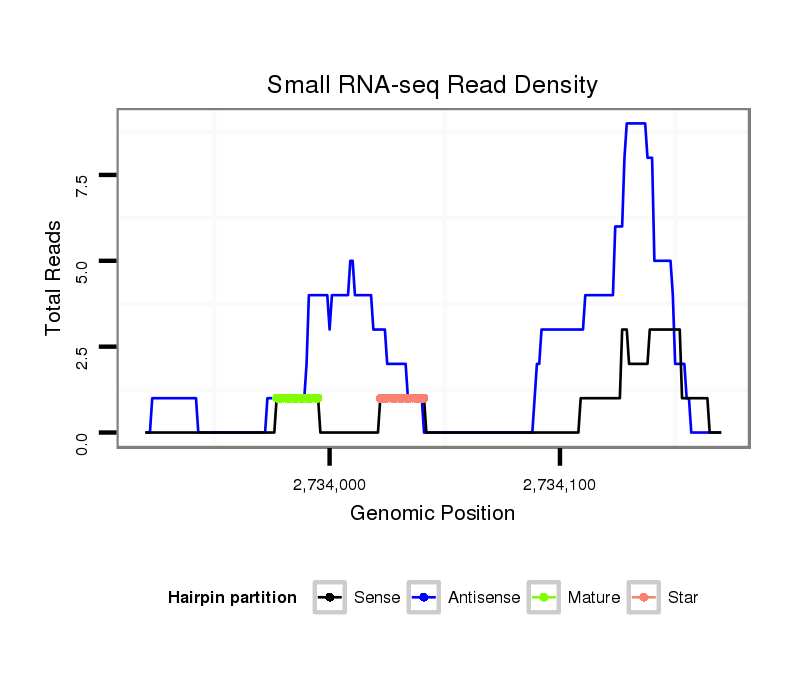
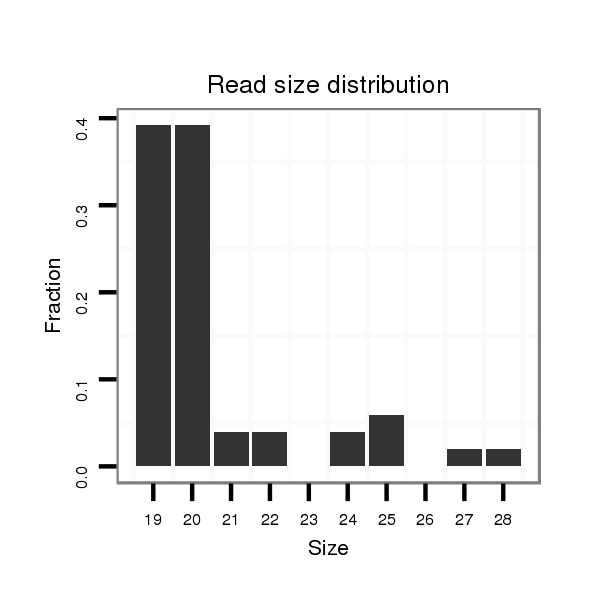
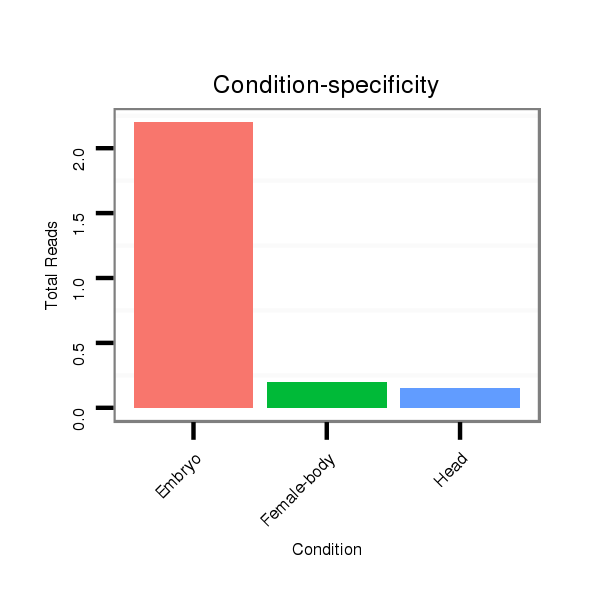
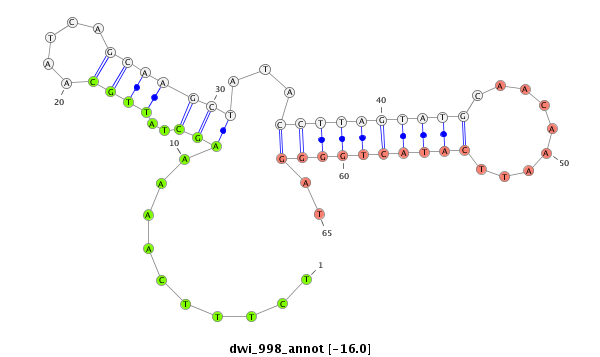
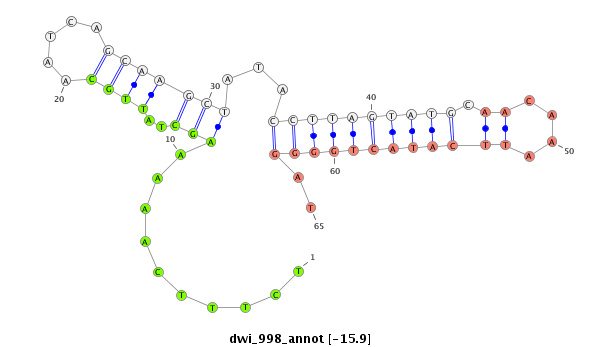
ID:dwi_998 |
Coordinate:scf2_1100000004514:2733970-2734120 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
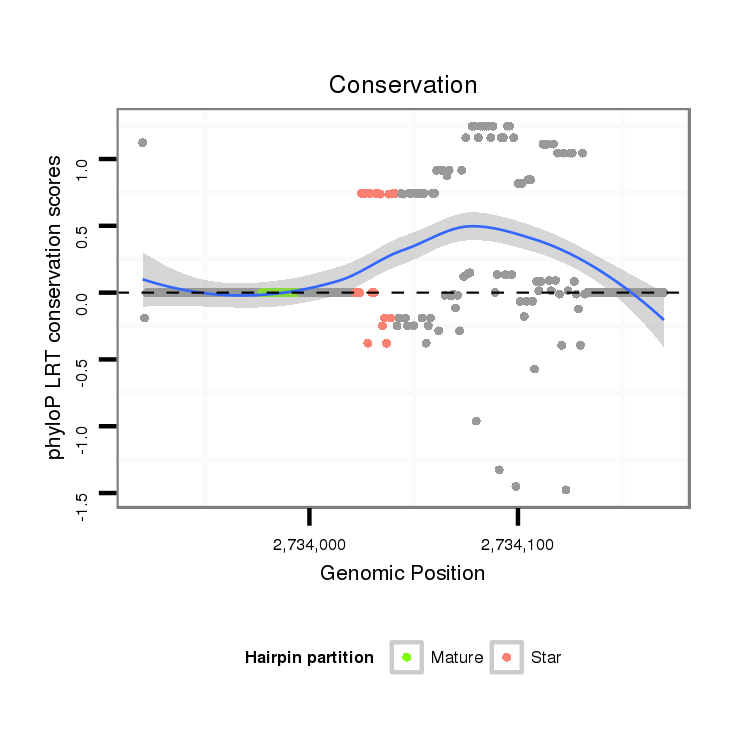
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -16.7 | -16.6 | -16.0 | -15.9 |
 |
 |
 |
 |
CDS [Dwil\GK15916-cds]; exon [dwil_GLEANR_16146:1]; intron [Dwil\GK15916-in]
| Name | Class | Family | Strand |
| Helitron-2N1_DVir | RC | Helitron | - |
| ---------#########################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATTTACACCATGAACGATCTATTTCATTTAAGTTTGGTATCATTTCGAAAGTAATGTTCTTTCAAAAAGCTATTGCAATCAGCAAGCTATACCTTAGTATGCAACAAATTCATACTGGGGATACTCCAATTTTTATGAAAATGTTTCTTCTAGAGCTATATGATATAGTGGTCCGATCCTGACAATTTTACTTTACCTGCTTCGGGCTAATGATGGTAATAATGGTACAAACGGGAAGAAAAGTTTGGGAG *********************************************************...............................((.((((((((((.........))))))))))))********************************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
V117 male body |
M045 female body |
V119 head |
M020 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................TAATGATGGTAATAATGGTACAAACG.................. | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................TCTTTCAAAAAGCTATTGC............................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................ACTTTACCTGCTTCGGGCTAA......................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................AACAAATTCATACTGGGGAT................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TAATGGTACAAACGGGAAGAAAAGTT...... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TAATAATGGTAATAATGGTACA...................... | 22 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TTCTGGATAAGGATGGTAAT............................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................TGATGGTAATAATAATACA...................... | 19 | 2 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................TATGATATAGTGGTCCGATCCTGAC.................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................TTCTAGAGCTATATGATATAGTGG................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................TGATATAGTGGTCCGATCCTGACAATTT............................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................TATAGTGGTCCGATCCTGACA................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................TCTAGAGCTATATGATATAGT.................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................ATTTTTATGAAAATGTTTCTTCTAGAG................................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................TATAGCTGAGTATGCAAGA................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................TAGTGGTCCGATCCTGACAATT................................................................ | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TAGAGCTATATGATATAGTGGTCCG............................................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................TATGAAAATGTTTCTTCTAGAG................................................................................................ | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................TATGAAAATGTTTCTTCTAGAGCT.............................................................................................. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................ATTTAAGTTTGCTATCGT................................................................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................GTTTCTTCTAGAGCTATATGATATA.................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
|
TAAATGTGGTACTTGCTAGATAAAGTAAATTCAAACCATAGTAAAGCTTTCATTACAAGAAAGTTTTTCGATAACGTTAGTCGTTCGATATGGAATCATACGTTGTTTAAGTATGACCCCTATGAGGTTAAAAATACTTTTACAAAGAAGATCTCGATATACTATATCACCAGGCTAGGACTGTTAAAATGAAATGGACGAAGCCCGATTACTACCATTATTACCATGTTTGCCCTTCTTTTCAAACCCTC
*********************************************************************************************************************************...............................((.((((((((((.........))))))))))))********************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V118 embryo |
M020 head |
V117 male body |
|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................GGACGAAGCCCGATTACTACCATTAT.............................. | 26 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................CCGATTACTACCATTATTACCATGTT..................... | 26 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 |
| ............................................................................................................................................................................GGCTAGGACTGTTAAAATGAAAT........................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................GGACTGTTAAAATGAAATGGATGAAG................................................ | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................TACTACCATTATTACCATGTTTGCCCTT.............. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................TTACTACCATTATTACCATGT...................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...ATGTGGTACTTGCTAGATAA.................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................GGACGAAGCCCGACTACTACCATTAT.............................. | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................AAATGGACGAAGCCCGATTACTACCAT................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................TACAAGAAAGTTTTTCGATAACGTTAG........................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................................CATACGTTGTTTAAGTATGACCCCT.................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................TAACGTTAGTCGTTCGATATGGAATCAT........................................................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................CGTTCGATATGGAATCATACGTTG.................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TGAAATGGACGAAGCCCGGTTACT...................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................CAGGCTAGGACTGTTAAAATGAAAT........................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................TAACGTTAGTCGTTCGATATGGAAT........................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................................ATGGAATCATACGTTGTTTAAGTAT......................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................ATAACGTTAGTCGTTCGATAT................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................CCAGGCTAGGACTGTTAAAATGAAAT........................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................TTACTACCATTATTACCATGTTTGCCC................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................TATCACCAGGCTAGGACTGTTAAAAT............................................................. | 26 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ......TGGGTCTTGCTAGATAAA................................................................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................TAAAATGACATGGACAAAG................................................ | 19 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................ATACTATATCACCAGGCTAGGAC...................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................CAAAGAAGATCTCGATATACTAT...................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................GATATACTATATCACCAGGCTAGGAC...................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004514:2733920-2734170 + | dwi_998 | ATTTACACCATGAACGATCTATTTCATTTAAGTTTGGTATCATTTCGAAAGTAATGTTCTTTCAAAAAGCTATTGCAATCAGCAAGCTATACCTTAGTATGCAACAAATTCATACTGGGGATACTCCAATTTTTATGAAAATGTTTCTTCTAGAGCTATATGATATAGTGGTCCGATCCTGACAATTTTACTTTACCTGCTTCGGGCTAATGATGGTAATAATGGTACAAACGGGAAGAAAAGTTTGGGAG |
| droAna3 | scaffold_13266:17032908-17032967 - | A---------------------------------------------------------------------------------------------------------------------------------------------------------GCTATAGGATATAGTCGACCGATCCCGGCCGTTCCGACTTATATACTGCCTGCAAAGGA-------------------------------------- | |
| droBip1 | scf7180000394982:9155-9214 + | A---------------------------------------------------------------------------------------------------------------------------------------------------------GCTATAGGATATAGTCGGCCGATCCGGGCCGTTCCGACTTATATACTGCGTGCAAAGGA-------------------------------------- | |
| droRho1 | scf7180000767788:65485-65547 - | ---------------------------------------------------------------------------------------------------------------------------------------------TGTTCCTATGGGATCTATACGATATAGTGAACCAATTC---------GGACTTATATACCACCTGCAGCAGG-------------------------------------- | |
| droTak1 | scf7180000415762:617026-617099 + | AC-------------------------------------------------------------------------------------------------------AAAGT--TACAATGAATTTTCTTATATTTGTTTGAATATTCCTATGGAAGCCTTAAGATATAGTGGTCCGATCC------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/18/2015 at 12:22 AM