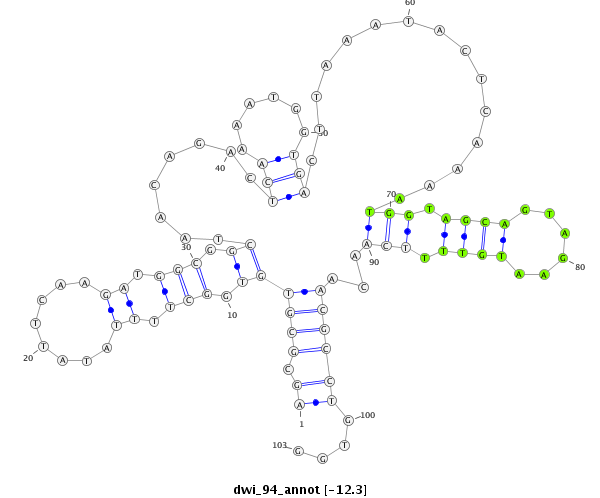
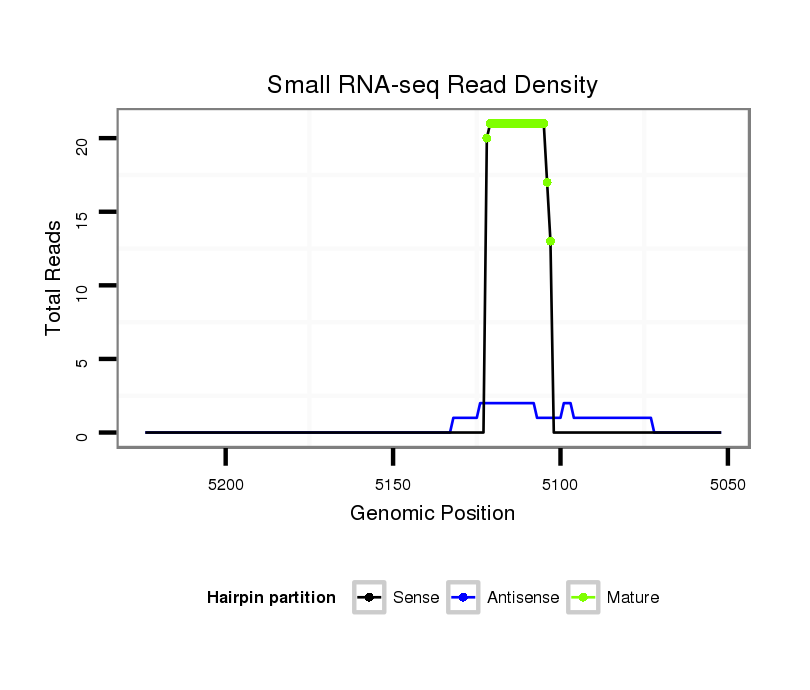
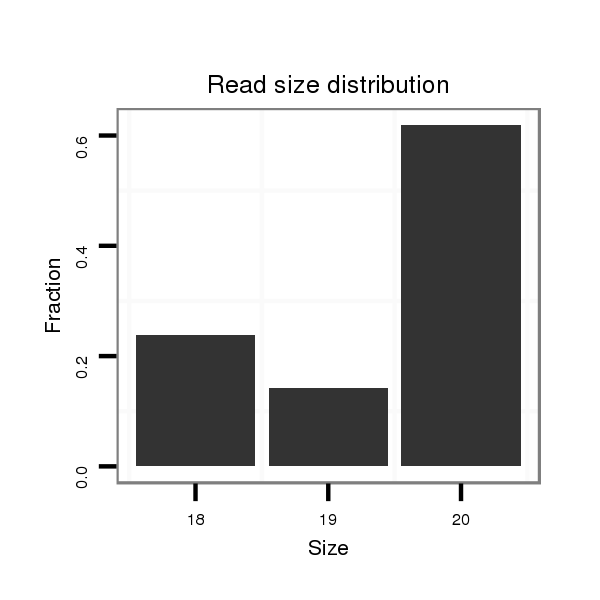
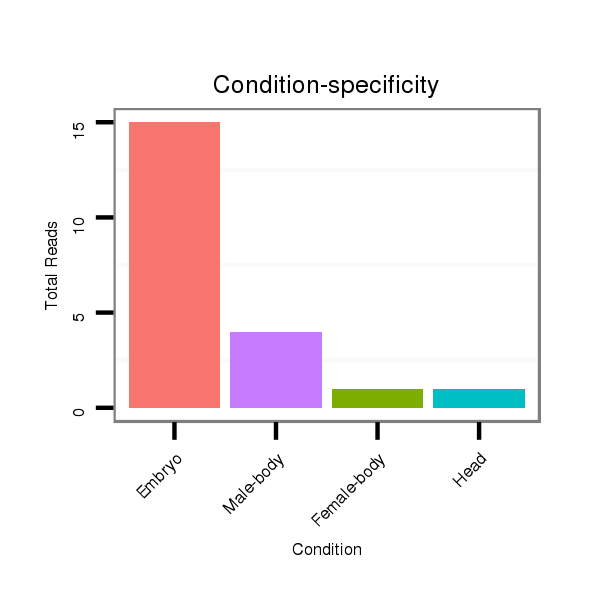
ID:dwi_94 |
Coordinate:scf2_1100000004943:5102-5174 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -12.2 | -12.0 | -11.9 |
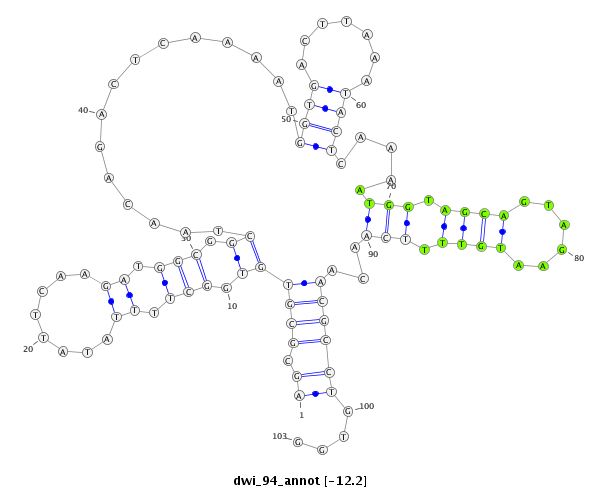 |
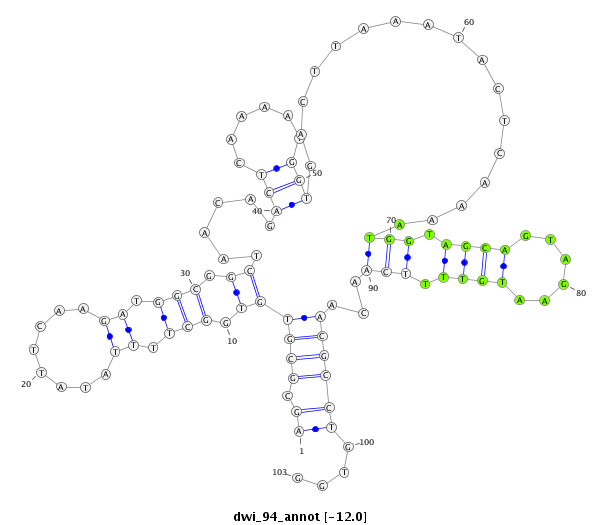 |
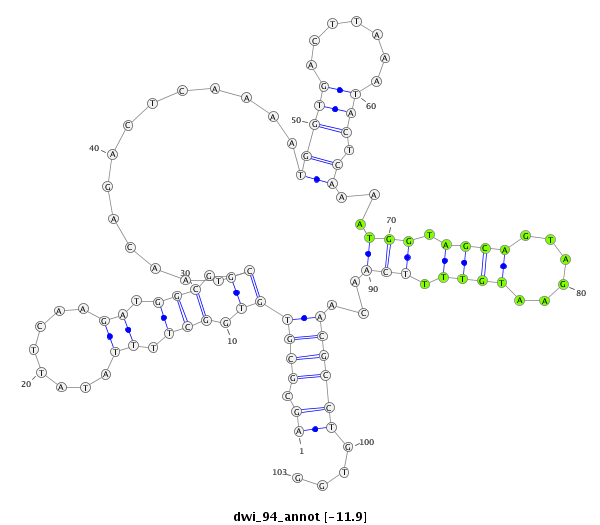 |
intergenic
No Repeatable elements found
|
AAGACTCAAAATGGTGGCTGTAGAATGGTTTCAGCAGCGCGTGTGGCTTTTATATTCAAGATGGCGGCTAACAGACTCAAAATGGTGACTTAAATACTCAAAATGGTAGCAGTAGAATGTTTTCAACAACGCCTGTGGCTTTTATATTAAAGATGGCGGCTAAAAAACTGAAA
***********************************((.((((((.(((.((........)).))).))........(((......)))...............(((.((((......)))).)))...))))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
V117 male body |
M045 female body |
V119 head |
|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................ATGGTAGCAGTAGAATGTTT................................................... | 20 | 0 | 1 | 13.00 | 13 | 10 | 3 | 0 | 0 |
| ......................................................................................................ATGGTAGCAGTAGAATGT..................................................... | 18 | 0 | 1 | 4.00 | 4 | 3 | 0 | 0 | 1 |
| ......................................................................................................ATGGTAGCAGTAGAATGTT.................................................... | 19 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 |
| .......................................................................................................TGGTAGCAGTAGAATGTT.................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ......................................................................................................ATGGTAGCAGTAGTATGTTT................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................TCAAAGTGGTAGCAGTAAAATGTT.................................................... | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ......................................................................................................ATGGAAGCAGTAGAATGTTT................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................CATGGTAGCAGTAGTATGTTT................................................... | 21 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................CATGGTAGCAGTAGAATGT..................................................... | 19 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ......................................................................................................ATGGTAGCGGTAAAATGTTTT.................................................. | 21 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................TGGCTTTTATATTCAAGATGGCGGC......................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
TTCTGAGTTTTACCACCGACATCTTACCAAAGTCGTCGCGCACACCGAAAATATAAGTTCTACCGCCGATTGTCTGAGTTTTACCACTGAATTTATGAGTTTTACCATCGTCATCTTACAAAAGTTGTTGCGGACACCGAAAATATAATTTCTACCGCCGATTTTTTGACTTT
***********************************((.((((((.(((.((........)).))).))........(((......)))...............(((.((((......)))).)))...))))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V118 embryo |
|---|---|---|---|---|---|---|---|
| ............................................................................................TTATGAGTTTTACCATCGTCATCTT........................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| .............................................................................................................................TGTTGCGGACACCGAAAATATAATTTC..................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| ....................................................................................................TTTACCATCGTCATCTTACAAAAGTTGT............................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ...................................................................................................................................................ATTTCTACCGCCGATTTTTTGACTTT | 26 | 0 | 2 | 0.50 | 1 | 1 | 0 |
| ......................................................AAGTTCTACCGCCGATTGTCTGAGTTT............................................................................................ | 27 | 0 | 9 | 0.11 | 1 | 1 | 0 |
| ....................................................ATAAGTTCTACCGCCGATTGTCTGAGT.............................................................................................. | 27 | 0 | 9 | 0.11 | 1 | 1 | 0 |
| .........................................................................CTGAGTTTTACCACTGAATTTATGAGT......................................................................... | 27 | 0 | 20 | 0.10 | 2 | 2 | 0 |
| ....................................CGCGCACACCGAAAATATAAGTTCT................................................................................................................ | 25 | 0 | 11 | 0.09 | 1 | 0 | 1 |
| ................................................AAATATAAGTTCTACCGCCGATTGT.................................................................................................... | 25 | 0 | 11 | 0.09 | 1 | 1 | 0 |
| .........................................ACACCGAAAATATAAGTTCTACCGCCGAT....................................................................................................... | 29 | 0 | 12 | 0.08 | 1 | 1 | 0 |
| ...........................................ACCGAAAATATAAGTTCTACCGCCGAT....................................................................................................... | 27 | 0 | 12 | 0.08 | 1 | 1 | 0 |
| ..................................................................CGATTGTCTGAGTTTTACCACTGAATT................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| .........................................................TTCTACCGCCGATTGTCTGAGTTT............................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| .....AGTTTTACCACCGACATCTTACCAAAGT............................................................................................................................................ | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| ...........................................................................GAGTTTTACCACTGAATTTAT............................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004943:5052-5224 - | dwi_94 | AAGACTCAAAATGGTGGCTGTAGAATGGTTTCAGCAGCGCGTGTGGCTTTTATATTCAAGATGGCGGCTAACAGACTCAAAATGGTGACTTAAATACTCAAAATGGTAGCAGTAGAATGTTTTCAACAACGCCTGTGGCTTTTATATTAAAGATGGCGGCTAAAAAACTGAAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 05/15/2015 at 02:35 PM