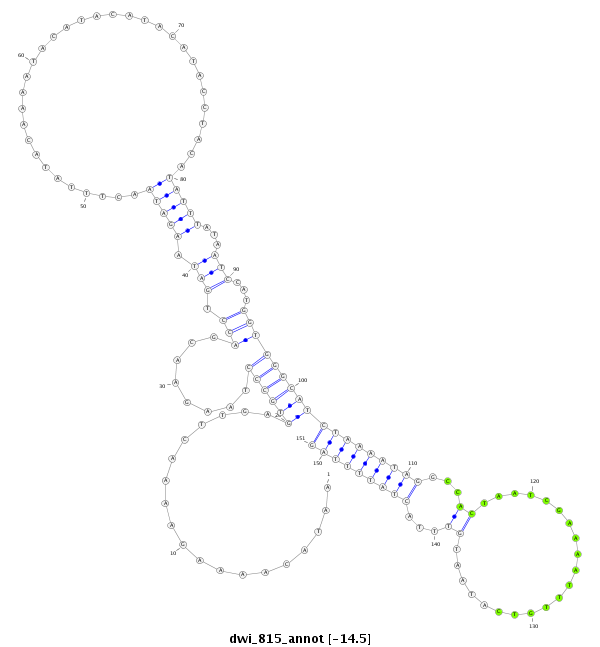
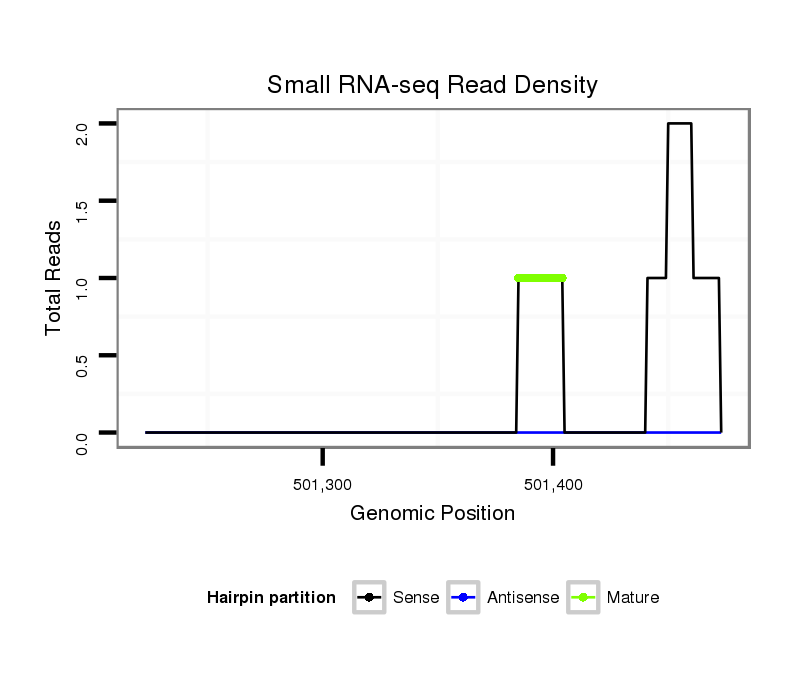
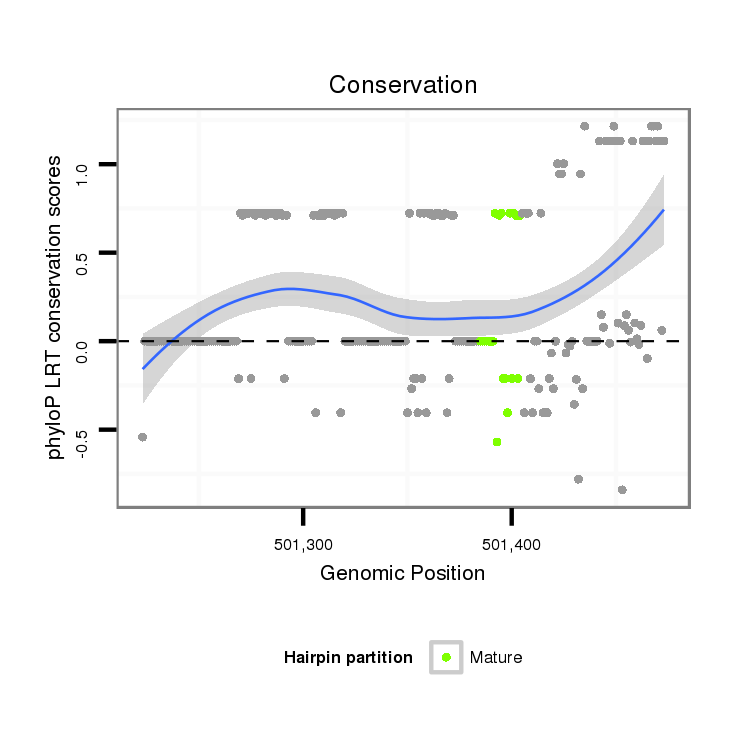
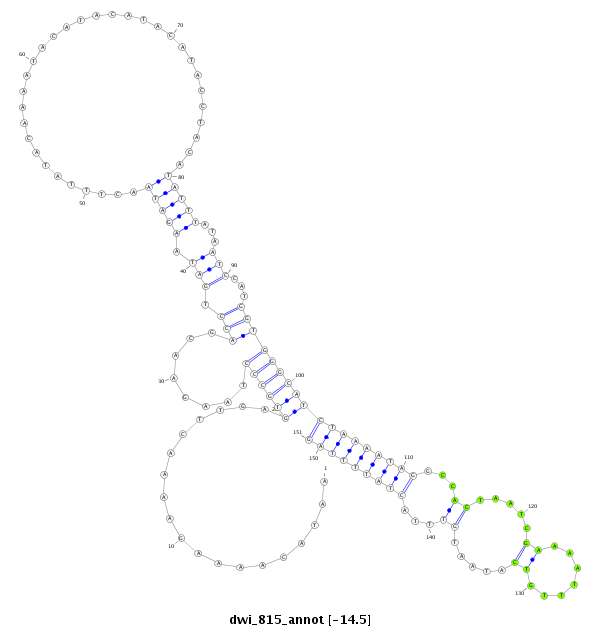
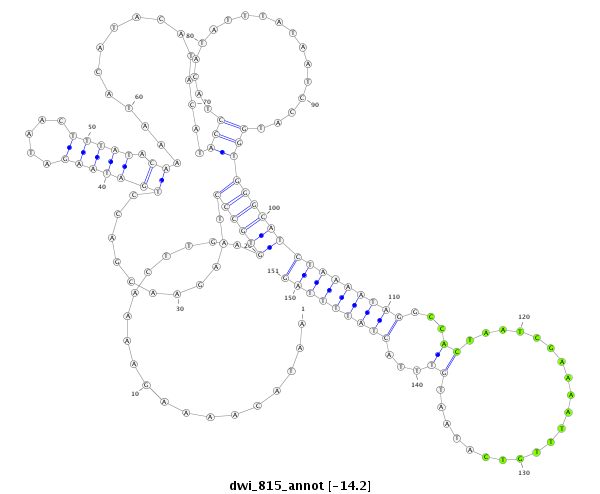
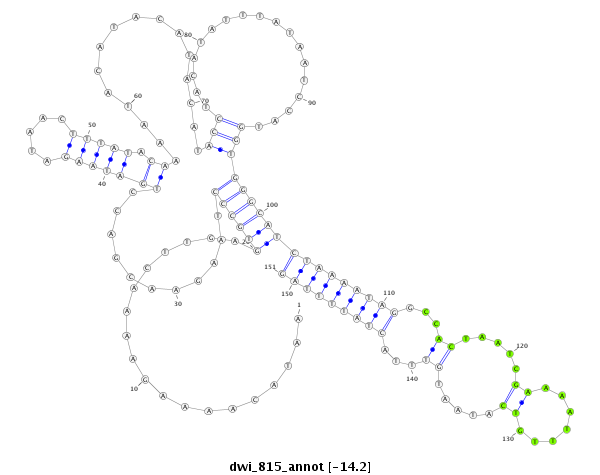
ID:dwi_815 |
Coordinate:scf2_1100000004513:501273-501423 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -14.5 | -14.2 | -14.2 |
 |
 |
 |
CDS [Dwil\GK21686-cds]; exon [dwil_GLEANR_4786:2]; intron [Dwil\GK21686-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CTCAAAGTAGACAAAAACAAATATGCCCTATACTAATTGTAAGTTATACAAATACAAAAGAAAACTTGAGTGCCCTAAGAACGACCTGATAAGATAACTTTATACAAAATACATACATACATACCTACATATTTATAATCCATGGTGGGCATCTAAAATAGGCCACTAATCGAAAATTTGTCATAATGTTTACTATTTTAGCAACTTCTACAAGCATCAGTTCCAGAGGCAAAGCCCCATCCAGATGAGCG **************************************************...................((((((........(((.(((.(((((.................................)))))...)))...)))))))))(((((((((...((.....................))...)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
M020 head |
M045 female body |
|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................AGTTCCAGAGGCAAAGCCCC............. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................................................................................................................................................CCACTAATCGAAAATTTGTC..................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................GGCAAAGCCCCATCCAGATGAGC. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................................AGTTATACAGATGCAAAAGAC............................................................................................................................................................................................. | 21 | 3 | 17 | 0.35 | 6 | 0 | 6 | 0 |
| .................................................................................................................................................................................................................................GAGGTAAAGGCCCATCAAGA...... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 |
| ............................................................................CAGCACAACCTGATAAGAT............................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ..........................................GTTATACAGATGCAAAAGAC............................................................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
|
GAGTTTCATCTGTTTTTGTTTATACGGGATATGATTAACATTCAATATGTTTATGTTTTCTTTTGAACTCACGGGATTCTTGCTGGACTATTCTATTGAAATATGTTTTATGTATGTATGTATGGATGTATAAATATTAGGTACCACCCGTAGATTTTATCCGGTGATTAGCTTTTAAACAGTATTACAAATGATAAAATCGTTGAAGATGTTCGTAGTCAAGGTCTCCGTTTCGGGGTAGGTCTACTCGC
**************************************************...................((((((........(((.(((.(((((.................................)))))...)))...)))))))))(((((((((...((.....................))...)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................GTCGGGCTTGGTCTACTCGC | 20 | 3 | 4 | 0.25 | 1 | 1 |
| ............................................................................................................................................................................................................................ATGGTCTCCGTTTCCGGGTG........... | 20 | 3 | 14 | 0.07 | 1 | 1 |
| ...............................................................................................................GTATGTATGTATGGATGTAT........................................................................................................................ | 20 | 0 | 16 | 0.06 | 1 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004513:501223-501473 + | dwi_815 | CTCAAAGTAGACAAAAACAAATATGCCCTATACTAATTGTAAGTTATACAAATACAAAAGAAAACT-----TGAGTGCCCTAAGAACGACCTGATAAGATAACTTTATACAAAATACATACATACATACCTACATATTTATAATCCATGGTGGGCATCTAAAATAGGCCACTAATCGAAAATTTGTCATAATGTTTACTAT----------------------------------------TTTAGCAACTTCTACAAGCATCAGTTCCAGAGGCAAAGCCCCATCCAGATGAGCG |
| dp5 | 3:11076026-11076075 + | T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCAGCAATACCAGCAA------GCCCCAGAGACCCGTGCAGAGCCAGATGAGAG | |
| droPer2 | scaffold_4:6394984-6395033 + | T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCAGCAATACCAGCAA------GCCCCAGAGACCCGTGCAGAGCCAGATGAGAG | |
| droKik1 | scf7180000302476:1399680-1399861 - | A---------------------------------------------CACAAACACAAAAGAAAACTACTCTTGGG------------GCCCTGATAAGATCA------------------------------AAAGCGTGTCATCCATGGTTAGC-------------------TGGAGGCTCTGCCAGAACT--AAAGCCTTTAGTAACGCACTTCTACGTATTTTTTCTCTTTCCAAATCGCAGCAGTACGTCCCA------GCTCCGGAGGCCCGGCCAGTGCCCGATGAGCG | |
| droTak1 | scf7180000415762:498935-498967 + | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A------GCTCCGGAGGCGCGGCCAGTGCCCGATGAGCG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/17/2015 at 11:55 PM