
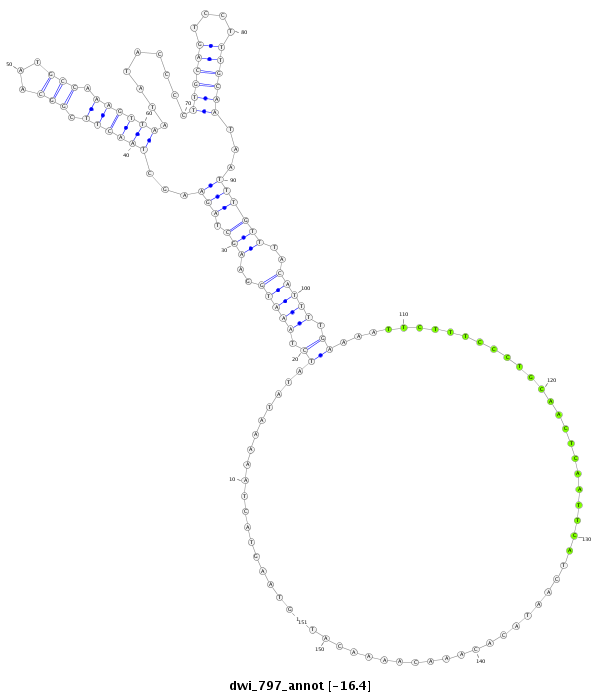
ID:dwi_797 |
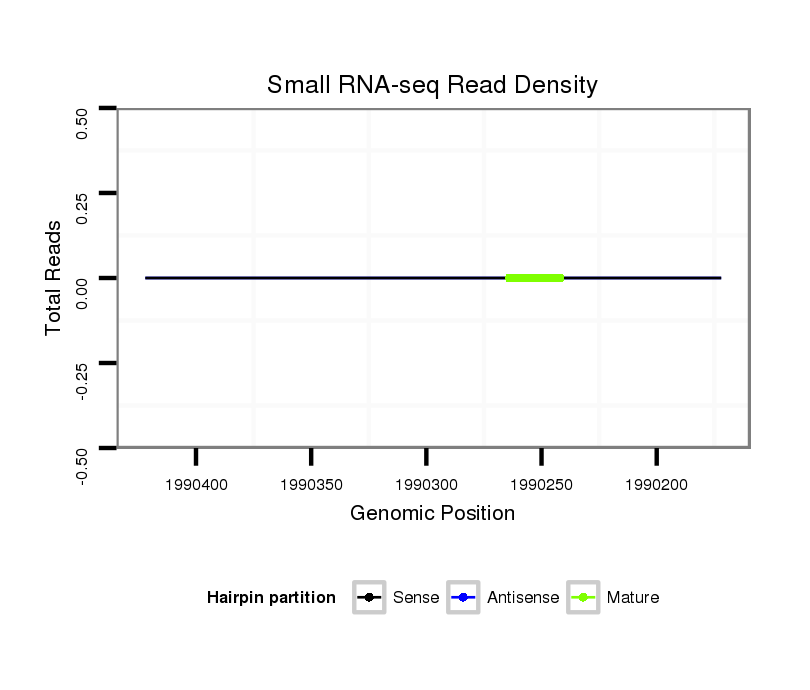
Coordinate:scf2_1100000004512:1990222-1990372 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -16.4 | -16.4 | -16.3 |
 |
 |
 |
CDS [Dwil\GK19478-cds]; exon [dwil_GLEANR_2109:1]; intron [Dwil\GK19478-in]
No Repeatable elements found
| --################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AAATGAAGAAGACTGAGGCAATGAAGCTTTTCCAAGAAGTAAAAGAAGTAGTAAGTACTAAAAATATATCTAAATGGAAGCTAGAAGCTAACTTCGGCAATGCCAAAGTTAATATACCCCTTGCAGTCCTTTGCAATAATTTGTTTACATTTTGAAAATTCTTTCCCTGCAACTCAATTCATCAATACACAAACAAAACATTAAAATTATTACTCTTTTTCCTACAATATTTTCTTCTTCTTCGATCATTC **************************************************.............................(((...)))((((((.(((...))).)))))).........((((((....))))))...(((((((.....((((..........................)))).....)))))))....************************************************** |
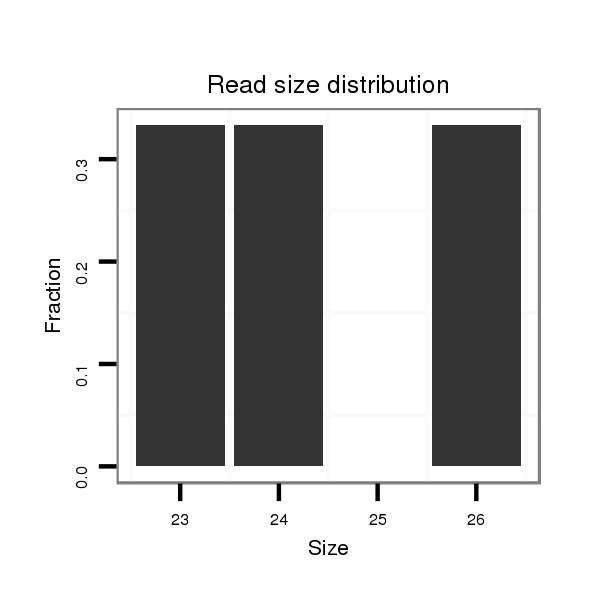
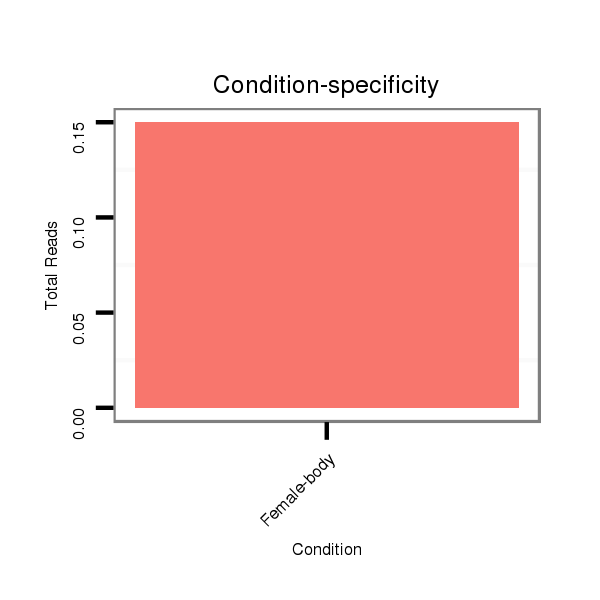
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
|---|---|---|---|---|---|---|
| ............................................................................TAAGCCAGAAGCTAACTTCGCC......................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 |
| ............................................................................................................................................................TATTCTTTCCCTGCAACTTAATTCATC.................................................................... | 27 | 2 | 19 | 0.05 | 1 | 1 |
| ..............................................................................................................................................................TTCTTTCCCTGCAACTCAATTCA...................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 |
| ...................................................................................................................................................................TCCCTGCAACTCAATTCATCAATACA.............................................................. | 26 | 0 | 20 | 0.05 | 1 | 1 |
| ...............................................................................................................................................................TCTTTCCCTGCAACTCAATTCATC.................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 |
|
TTTACTTCTTCTGACTCCGTTACTTCGAAAAGGTTCTTCATTTTCTTCATCATTCATGATTTTTATATAGATTTACCTTCGATCTTCGATTGAAGCCGTTACGGTTTCAATTATATGGGGAACGTCAGGAAACGTTATTAAACAAATGTAAAACTTTTAAGAAAGGGACGTTGAGTTAAGTAGTTATGTGTTTGTTTTGTAATTTTAATAATGAGAAAAAGGATGTTATAAAAGAAGAAGAAGCTAGTAAG
**************************************************.............................(((...)))((((((.(((...))).)))))).........((((((....))))))...(((((((.....((((..........................)))).....)))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V118 embryo |
V117 male body |
|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................AGGGACGTTGAGTTAAGTAGTTAT................................................................ | 24 | 0 | 20 | 0.55 | 11 | 11 | 0 | 0 |
| ..................................................................................................................................................................AAGGGACGTTGAGTTAAGTAGTTAT................................................................ | 25 | 0 | 20 | 0.40 | 8 | 6 | 0 | 2 |
| .....................................................................................................................................................................GGACGTTGAGTTAAGTAGTTAT................................................................ | 22 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 |
| .................................................................................................................................................................AAAGGGACGTTGAGTTAAGTAGTTATG............................................................... | 27 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 |
| ................................................................................................................................................................GAAAGGGACGTTGAGTTAAGTA..................................................................... | 22 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 |
| ..................................................................................................................................................................................AGTAGTTATGTGTTTGTTTTGTAAT................................................ | 25 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 |
| .........................................................................................................................................................................GTTGAGTTAAGTAGTTAT................................................................ | 18 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 |
| ................................................................................................CGTTCCGGTTTCAATGACAT....................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 |
| .....................................................................................................................................................................................AGTTATGTGTTTGTTTTG.................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ..................................................................................................................................................................AAGGGACGTTGAGTTAAGTAGTT.................................................................. | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ..............................................................................................................................................................................GTTAAGTAGTTATGTGTTTGTTTTGT................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ..................................................................................................................................................................AAGGGACGTTGAGTTAAGTA..................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| .........................................................................................................................................................................GTTGAGTTAAGTAGTTATG............................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ..................................................................................TCTTCGATTGAAGCCGTTACG.................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ..........................................................................................................................................................................TTGAGTTAAGTAGTTATGTGTTTGT........................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................................GAAAGGGACGTTGAGTTAAGTAGTTAT................................................................ | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................................AGGGACGTTGAGTTAAGTAG.................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ..................................................................................................................................................................AAGGTACGTTGAGTTAAGTAGTTAT................................................................ | 25 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ....................................................................................................................................................................GGGACGTTGAGTTAAGTAG.................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004512:1990172-1990422 - | dwi_797 | AAATGAAGAAGACTGAGGCAATGAAGCTTTTCCAAGAAGTAAAAGAAGTAGTAAGTACTAAAAATATATCTAAATGGAAGCTAGAAGCTAACTTCGGCAATGCCAAAGTTAATATACCCCTTGCAGTCCTTTGCAATAATTTGTTTACATTTTGAAAATTCTTTCCCTGCAACTCAATTCATCAATACACAAACAAAACATTAAAATTATTACTCTTTTTCCTACAATATTTTCTTCTTCTTCGATCATTC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 05/17/2015 at 11:57 PM