
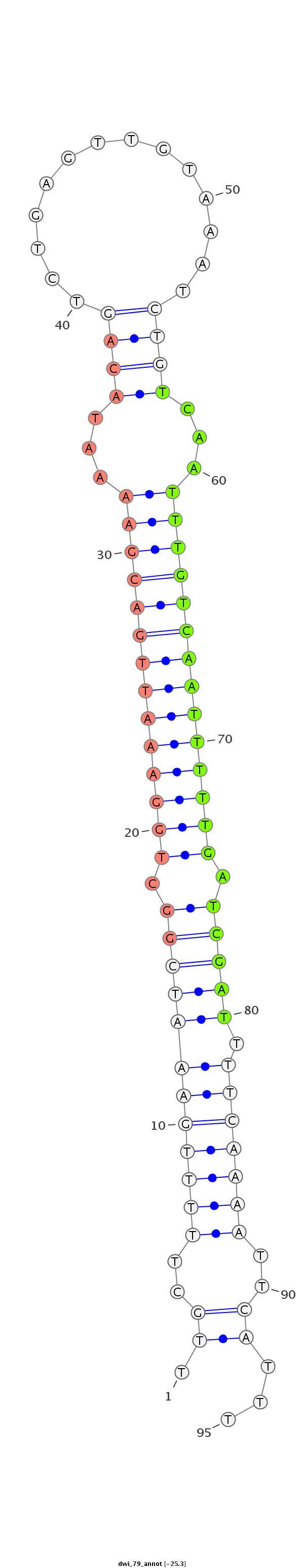
ID:dwi_79 |
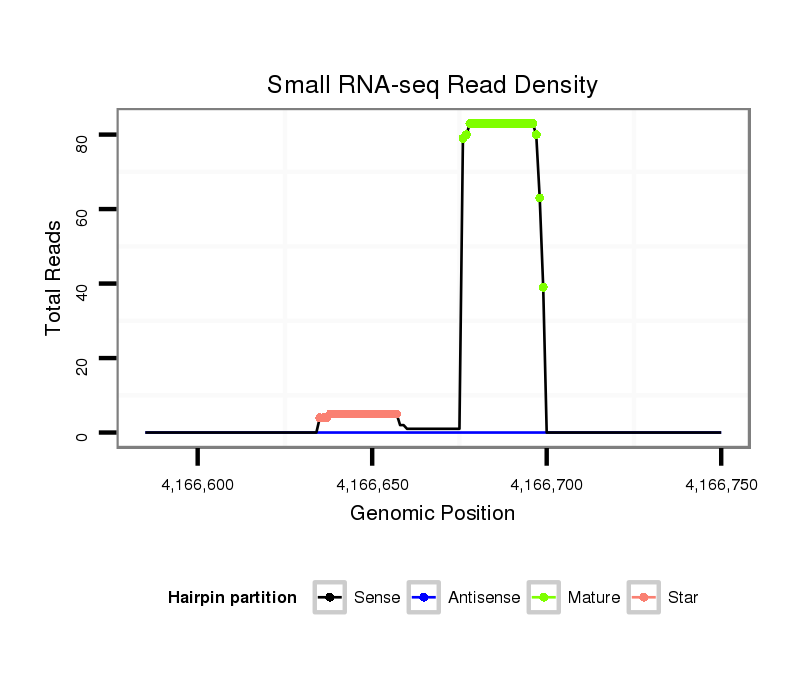
Coordinate:scf2_1100000004909:4166635-4166700 + |
Confidence:confident |
Class:Canonical miRNA |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -25.3 | -25.3 | -25.3 |
 |
 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
TTTTTTTTTTTTAAGTCGTCTTGAAATTAATTAATTTGCTTTTTGAAATCGGCTGGAAATTGACGAAAATACAGTCTGAGTTGTAAATCTGTCAATTTGTCAATTTTTGATCGATTTTCAAAATTCATTTCCACCAAGAAGTAAAATTTTGACCATTTTCGCATAA
***********************************.((..((((((((((((.((((((((((((((...((((..............))))...)))))))))))))).)))).))))))))..))...************************************ |
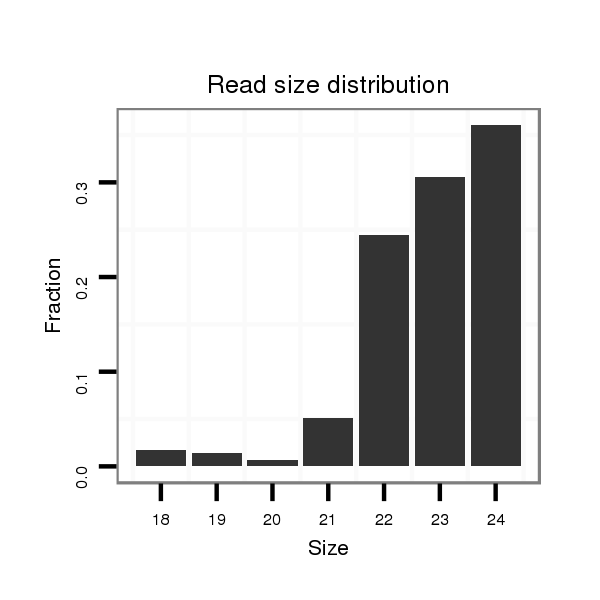
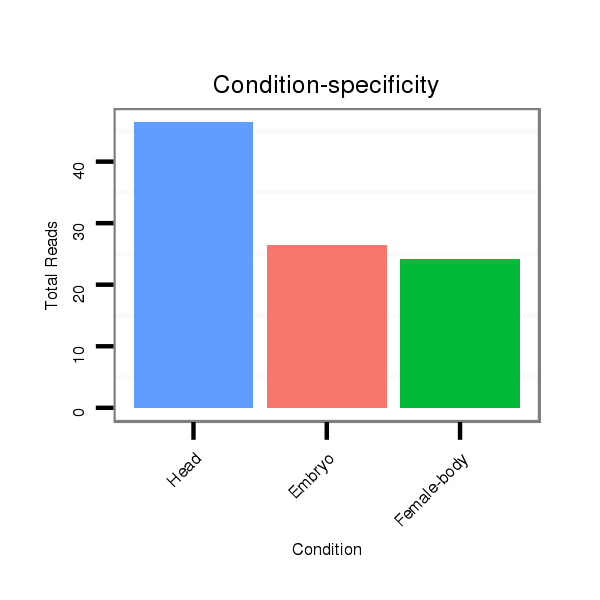
Read size | # Mismatch | Hit Count | Total Norm | Total | M020 head |
V118 embryo |
M045 female body |
V119 head |
V117 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................TCAATTTGTCAATTTTTGATCGAT................................................... | 24 | 0 | 3 | 35.00 | 105 | 39 | 33 | 10 | 23 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGATCGA.................................................... | 23 | 0 | 3 | 24.00 | 72 | 24 | 22 | 13 | 13 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGATCG..................................................... | 22 | 0 | 3 | 17.33 | 52 | 13 | 3 | 30 | 6 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGATCGT.................................................... | 23 | 1 | 3 | 8.67 | 26 | 26 | 0 | 0 | 0 | 0 |
| ..................................................GGCTGGAAATTGACGAAAATACA............................................................................................. | 23 | 0 | 3 | 4.33 | 13 | 0 | 11 | 1 | 1 | 0 |
| .............................................................................................AATTTGTCAATTTTTGATCGAT................................................... | 22 | 0 | 3 | 3.67 | 11 | 6 | 0 | 2 | 3 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGATC...................................................... | 21 | 0 | 3 | 3.33 | 10 | 0 | 0 | 5 | 5 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGATCGG.................................................... | 23 | 1 | 3 | 1.67 | 5 | 5 | 0 | 0 | 0 | 0 |
| .....................................................TGGAAATTGACGAAAATACAGT........................................................................................... | 22 | 0 | 5 | 1.40 | 7 | 0 | 4 | 1 | 2 | 0 |
| .........................................................................GTCTGAGTTGTAAATCTG........................................................................... | 18 | 0 | 3 | 1.33 | 4 | 0 | 4 | 0 | 0 | 0 |
| ............................................................................................CAATTTGTCAATTTTTGATCGAT................................................... | 23 | 0 | 3 | 1.33 | 4 | 2 | 0 | 2 | 0 | 0 |
| .............................................................................................AATTTGTCAATTTTTGATCGA.................................................... | 21 | 0 | 3 | 0.67 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGA........................................................ | 19 | 0 | 3 | 0.67 | 2 | 0 | 0 | 2 | 0 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGATCGAC................................................... | 24 | 1 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................TGTCAATTTGTCAATTTTTGA........................................................ | 21 | 0 | 3 | 0.67 | 2 | 0 | 0 | 1 | 1 | 0 |
| ...........................................................................................TCAATTTTTCAATTTTTGATCGAT................................................... | 24 | 1 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...................................................GCTGGAAATTGACGAAAATACA............................................................................................. | 22 | 0 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 |
| ....................................................................................................................TTCTACATTCATTACCACCA.............................. | 20 | 3 | 20 | 0.40 | 8 | 0 | 0 | 6 | 2 | 0 |
| .........................................................................................TGTCAATTTGTCAATTTTTGATT...................................................... | 23 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................CAATTTGTCAATTTTTGATC...................................................... | 20 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................GTCAATTTGTCAATTTTTG......................................................... | 19 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................TGTCAATTTGTCAATTTTTGAT....................................................... | 22 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGATCAAT................................................... | 24 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................TCAATTTCTCAATTTTTGATCGAT................................................... | 24 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................TTCAATTTGTCAATTTTTGATCG..................................................... | 23 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGATAG..................................................... | 22 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................TTTGTCAATTTTTGATCG..................................................... | 18 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................TCAATTTGTCAGTTTTTGATCGA.................................................... | 23 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGATCGCT................................................... | 24 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................CAATTTGTCAATTTTTGATCG..................................................... | 21 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................TTAATTTGTCAATTTTTGATCGAT................................................... | 24 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................TCAATTTGTCAATTTTTGAT....................................................... | 20 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................CCAATTTGTCAATTTTTGATCGAT................................................... | 24 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................GGCTGGAAATTGACGAAAATAC.............................................................................................. | 22 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................TTGTCAATTTTTGATCGAT................................................... | 19 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................TCAATTTGTTAATTTTTGATCGAT................................................... | 24 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................TGGAAATTGACGAACATACAGT........................................................................................... | 22 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................................................TGGAAATTGAAGAAAATACAGT........................................................................................... | 22 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 |
|
AAAAAAAAAAAATTCAGCAGAACTTTAATTAATTAAACGAAAAACTTTAGCCGACCTTTAACTGCTTTTATGTCAGACTCAACATTTAGACAGTTAAACAGTTAAAAACTAGCTAAAAGTTTTAAGTAAAGGTGGTTCTTCATTTTAAAACTGGTAAAAGCGTATT
************************************.((..((((((((((((.((((((((((((((...((((..............))))...)))))))))))))).)))).))))))))..))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
|---|---|---|---|---|---|---|
| ..AAAAAAAAAATTCAGCAGGAC............................................................................................................................................... | 21 | 1 | 4 | 0.25 | 1 | 1 |
| ..AAAAAAAAAATTCAGCAG.................................................................................................................................................. | 18 | 0 | 4 | 0.25 | 1 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droWil2 | scf2_1100000004909:4166585-4166750 + | dwi_79 | confident | TTTTTTTTTTTTAAGTCGTCTTGAAATTAATTAATTTGCTTTTTGAAATCGGCTGGAAATTGACGAAAATACAGTCTGAGTTGTAAATCTGTCAATTTGTCAATTTTTGATCGATTTTCAAAATTCATTTCCACCAAGAAGTAAAATTTTGACCATTTTCGCATAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 10/20/2015 at 07:04 PM