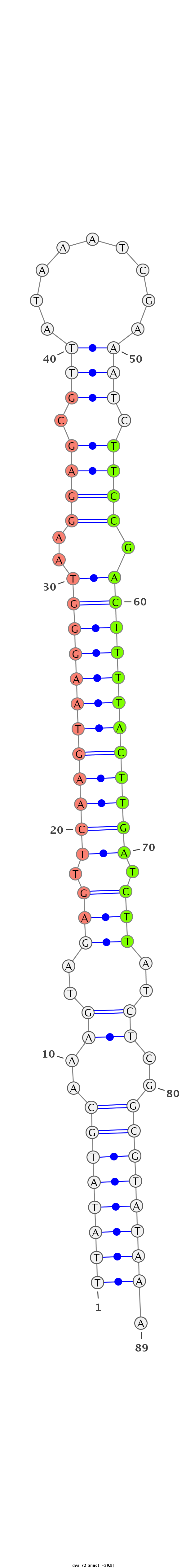
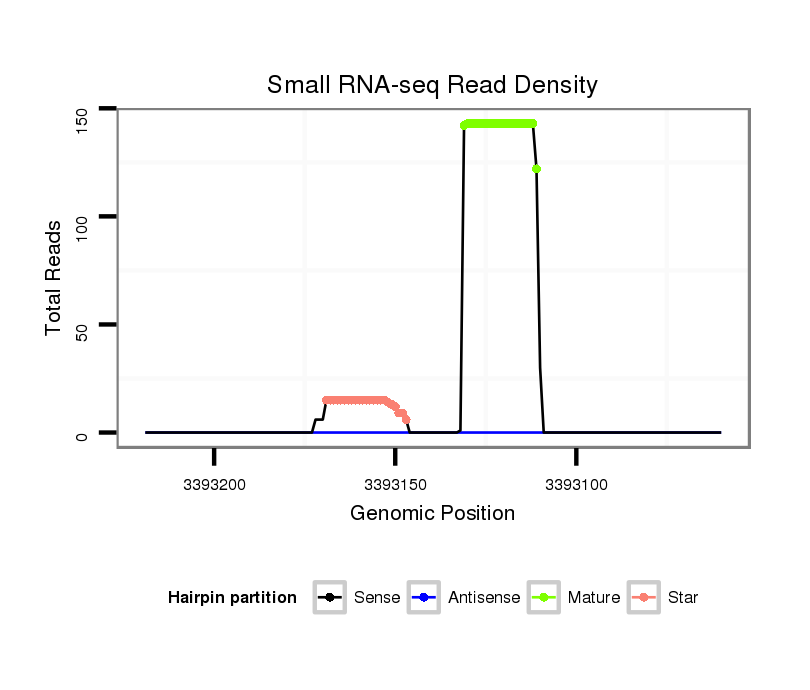
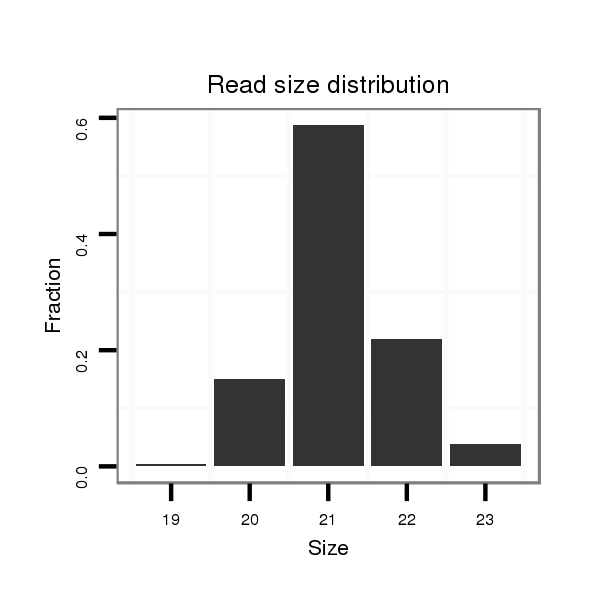
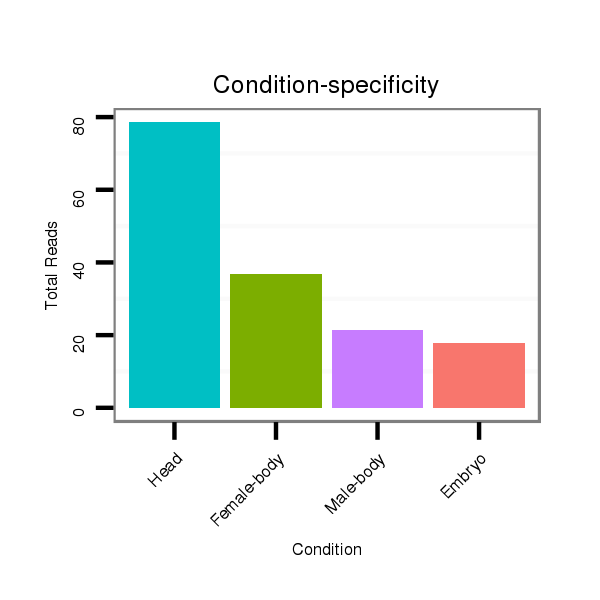
ID:dwi_72 |
Coordinate:scf2_1100000004511:3393110-3393169 - |
Confidence:confident |
Class:Testes-restricted |
Genomic Locale:antisense_to_intron |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -29.8 | -29.4 | -29.0 |
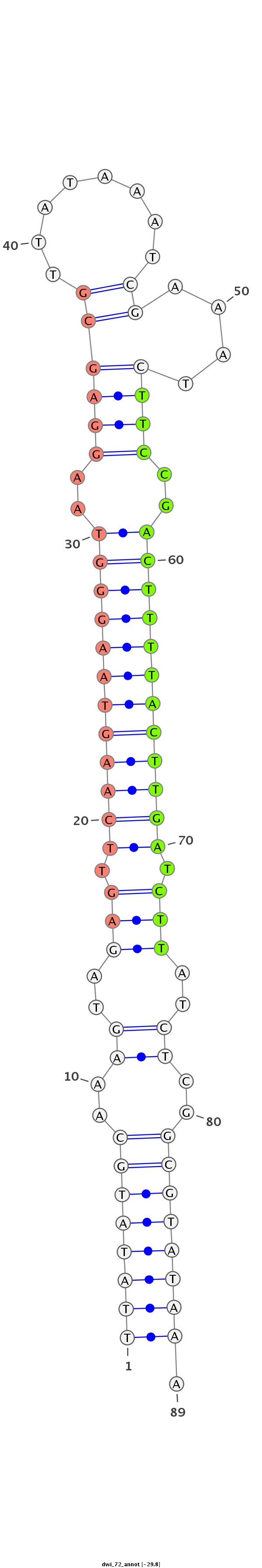 |
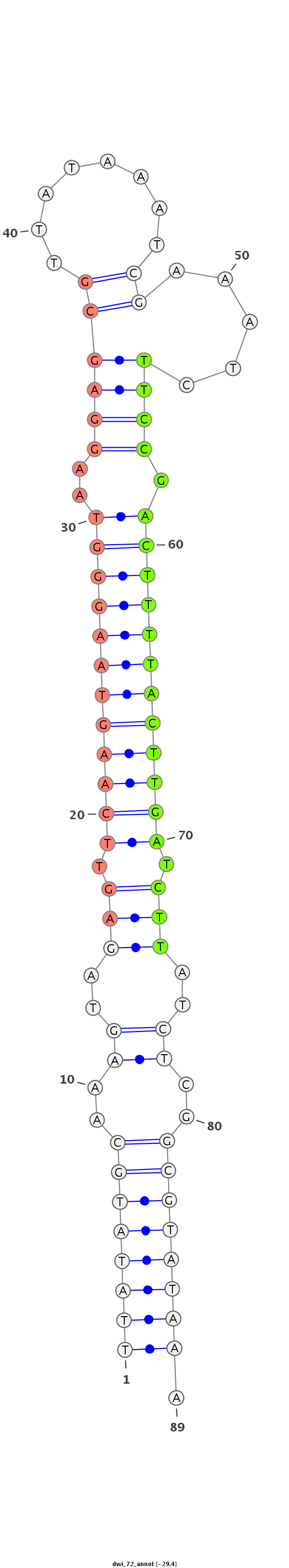 |
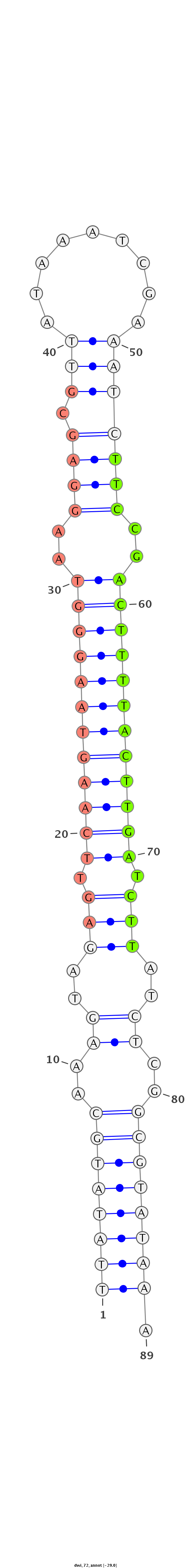 |
Antisense to intron [Dwil\GK17265-in]
No Repeatable elements found
| mature | star |
|
CTAAGTGATCGTCGCAGGATCATAAAAATCATATCTTATATGCAAAGTAGAGTTCAAGTAAGGGTAAGGAGCGTTATAAATCGAAATCTTCCGACTTTTACTTGATCTTATCTCGGCGTATAAAAAACTCATAAAAAAAATACAAATCAACATTTTTCTA
***********************************((((((((..((..(((.((((((((((((..((((.(((.........))).)))).)))))))))))).)))..))..)))))))).************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M020 head |
M045 female body |
V117 male body |
V118 embryo |
V119 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................TTCCGACTTTTACTTGATCTT................................................... | 21 | 0 | 3 | 90.00 | 270 | 165 | 62 | 24 | 11 | 8 |
| ........................................................................................TTCCGACTTTTACTTGATCTTA.................................................. | 22 | 0 | 3 | 30.00 | 90 | 5 | 35 | 15 | 35 | 0 |
| ........................................................................................TTCCGACTTTTACTTGATCT.................................................... | 20 | 0 | 3 | 21.00 | 63 | 55 | 3 | 4 | 0 | 1 |
| ..................................................AGTTCAAGTAAGGGTAAGGAGCG....................................................................................... | 23 | 0 | 4 | 6.00 | 24 | 0 | 3 | 15 | 6 | 0 |
| ........................................................................................TTCCGACTTTTACTTGATCTTAA................................................. | 23 | 1 | 3 | 4.67 | 14 | 2 | 10 | 0 | 1 | 1 |
| ...............................................TAGAGTTCAAGTAAGGGTAAGGA.......................................................................................... | 23 | 0 | 4 | 3.50 | 14 | 0 | 0 | 5 | 9 | 0 |
| ..................................................AGTTCAAGTAAGGGTAAGGAGC........................................................................................ | 22 | 0 | 4 | 3.00 | 12 | 0 | 2 | 9 | 0 | 1 |
| ...............................................TAGAGTTCAAGTAAGGGTAAG............................................................................................ | 21 | 0 | 4 | 1.75 | 7 | 0 | 0 | 7 | 0 | 0 |
| ...............................................TAGAGTTCAAGTAAGGGTAAGG........................................................................................... | 22 | 0 | 4 | 1.75 | 7 | 0 | 0 | 6 | 1 | 0 |
| .........................................................................................TCCGACTTTTACTTGATCTT................................................... | 20 | 0 | 3 | 1.67 | 5 | 0 | 5 | 0 | 0 | 0 |
| ...............................................TAGAGTTCAAGTAAGGGTAA............................................................................................. | 20 | 0 | 4 | 1.00 | 4 | 0 | 0 | 3 | 1 | 0 |
| ......................................................................................GCTTCCGACTTTTACTTGATCTT................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................TTCCGACTTTTAATTGATCTT................................................... | 21 | 1 | 3 | 1.00 | 3 | 2 | 0 | 0 | 1 | 0 |
| .......................................................................................CTTCCGACTTTTACTTGATCTT................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................TTCCGACTTTTACTTGATCTTC.................................................. | 22 | 1 | 3 | 0.67 | 2 | 1 | 1 | 0 | 0 | 0 |
| ..................................................AGTTCAAGTAAGGGTAAGGA.......................................................................................... | 20 | 0 | 4 | 0.50 | 2 | 0 | 0 | 2 | 0 | 0 |
| ..................................................AGTTCAAGTAAGGGTAAGGAG......................................................................................... | 21 | 0 | 4 | 0.50 | 2 | 0 | 1 | 1 | 0 | 0 |
| ........................................................................................TGCCGACTTTTACTTGATCTT................................................... | 21 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................TTCCGACTTTTGCTTGATCT.................................................... | 20 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................TTCCGACTTTTCCTTGATCTTAA................................................. | 23 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................CGACTTTTACTTGATCTTATCTA.............................................. | 23 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................TTCCGACTTTTACTTGATCTTT.................................................. | 22 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................TTCCGACTTTTACTTGATC..................................................... | 19 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................TTCCGACTTTTACTTGATCTC................................................... | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TCCGACTTTTACTTGATCTTA.................................................. | 21 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................TTCCGACTTTTACTTGATCC.................................................... | 20 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TTCCGACTTTTACTTGATTTT................................................... | 21 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................TTCCGACCTTTACTTGATCTTC.................................................. | 22 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................TTCCGACTTTTACTTGATCG.................................................... | 20 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TCCGACTTTTACTTGATCTTAA................................................. | 22 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................AGTTCAAGTAAGGGTAAGG........................................................................................... | 19 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................TAGAGTTCAAGTAAGGGTAAGGAA......................................................................................... | 24 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................TAGGGTTCAAGTAAGGGTAAGG........................................................................................... | 22 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................TAGAGTTCCAGTAAGGGTAAG............................................................................................ | 21 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................AGTTCAACTAAGGGTAAGGAGCG....................................................................................... | 23 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................TAGAGTTCAAGTAAGGTTAAGGAG......................................................................................... | 24 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................GGTTCAAGTAAGGGTAAGGAGCG....................................................................................... | 23 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................AGAGTTCAAGTAAGGGTAAG............................................................................................ | 20 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................TAGAGTTCAAGTAAGCGTAAGGAG......................................................................................... | 24 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................TAGAGTTCAAGTAAGCGTAAGGA.......................................................................................... | 23 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................TAGAGTTCAAGTAAGGGTAAAG........................................................................................... | 22 | 1 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................TAGAGTTCAAGTAAGGGTAAAGA.......................................................................................... | 23 | 1 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................TAGAGTTCAAGTAAGGGTAAAGAA......................................................................................... | 24 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...............................................TAGAGTTCAAGTAAGGGTAAA............................................................................................ | 21 | 1 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 |
|
GATTCACTAGCAGCGTCCTAGTATTTTTAGTATAGAATATACGTTTCATCTCAAGTTCATTCCCATTCCTCGCAATATTTAGCTTTAGAAGGCTGAAAATGAACTAGAATAGAGCCGCATATTTTTTGAGTATTTTTTTTATGTTTAGTTGTAAAAAGAT
************************************((((((((..((..(((.((((((((((((..((((.(((.........))).)))).)))))))))))).)))..))..)))))))).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V117 male body |
V118 embryo |
|---|---|---|---|---|---|---|---|
| .................................................................................CCTTTGGAATGCTGAAAATGA.......................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 |
| ........................................................................................................................ATTTCTCGAGTATTTTTTTTATAT................ | 24 | 3 | 6 | 0.17 | 1 | 0 | 1 |
| ...................................................................................TTTGGAATGCTGAAAATG........................................................... | 18 | 2 | 19 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droWil2 | scf2_1100000004511:3393060-3393219 - | dwi_72 | confident | CTAAGTGATCGTCGCAGGATCATAAAAATCATATCTTATATGCAAAGTAGAGTTCAAGTAAGGGTAAGGAGCGTTATAAATCGAAATCTTCCGACTTTTACTTGATCTTATCTCGGCGTATAAAAAACTCATAAAAAAAATACAAATCAACATTTTTCTA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 10/20/2015 at 07:36 PM