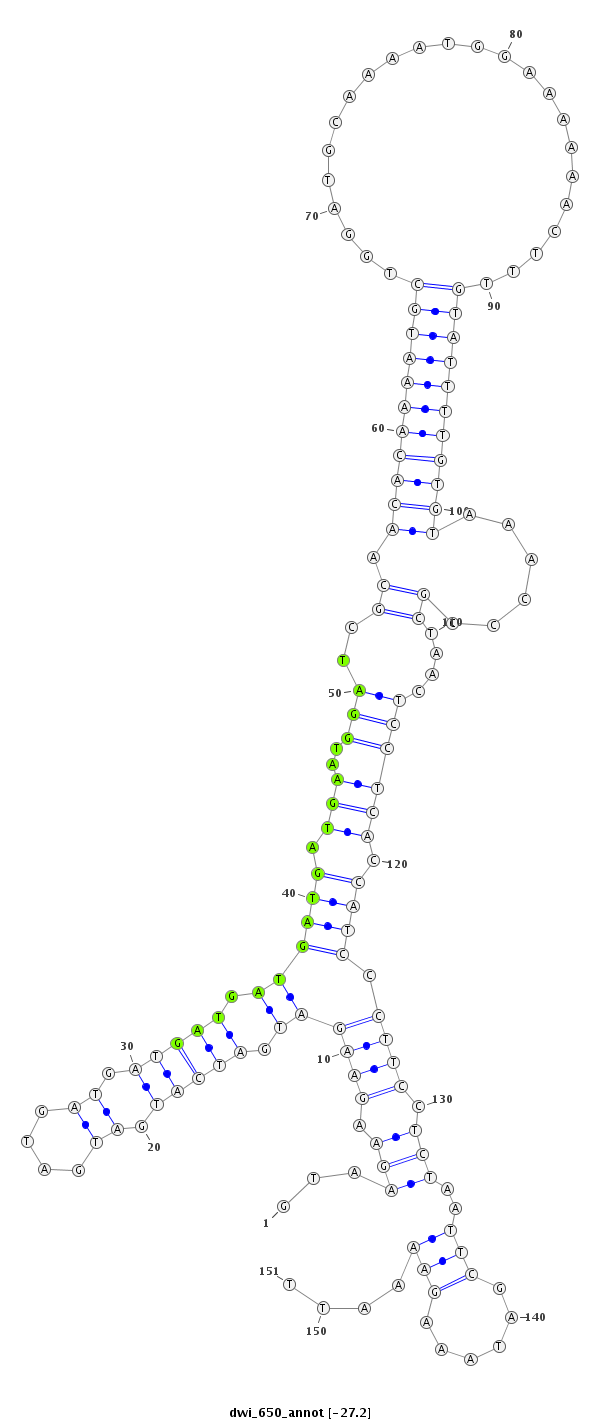
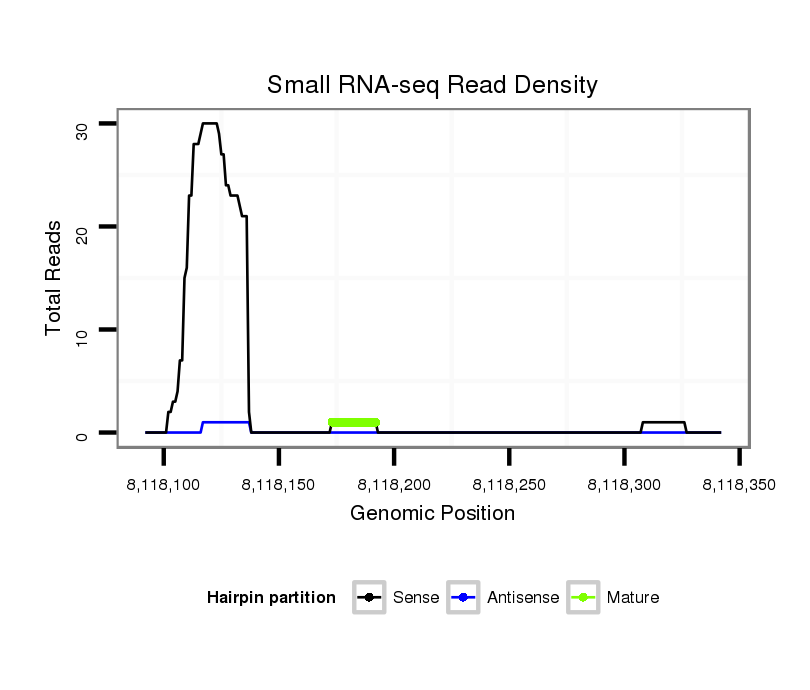
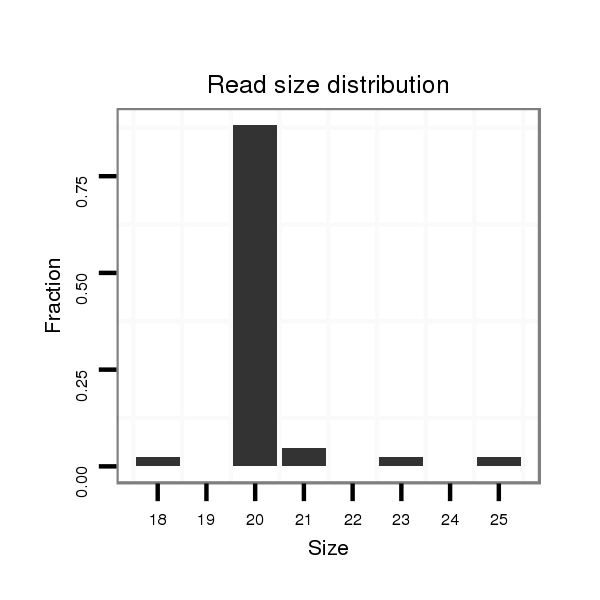
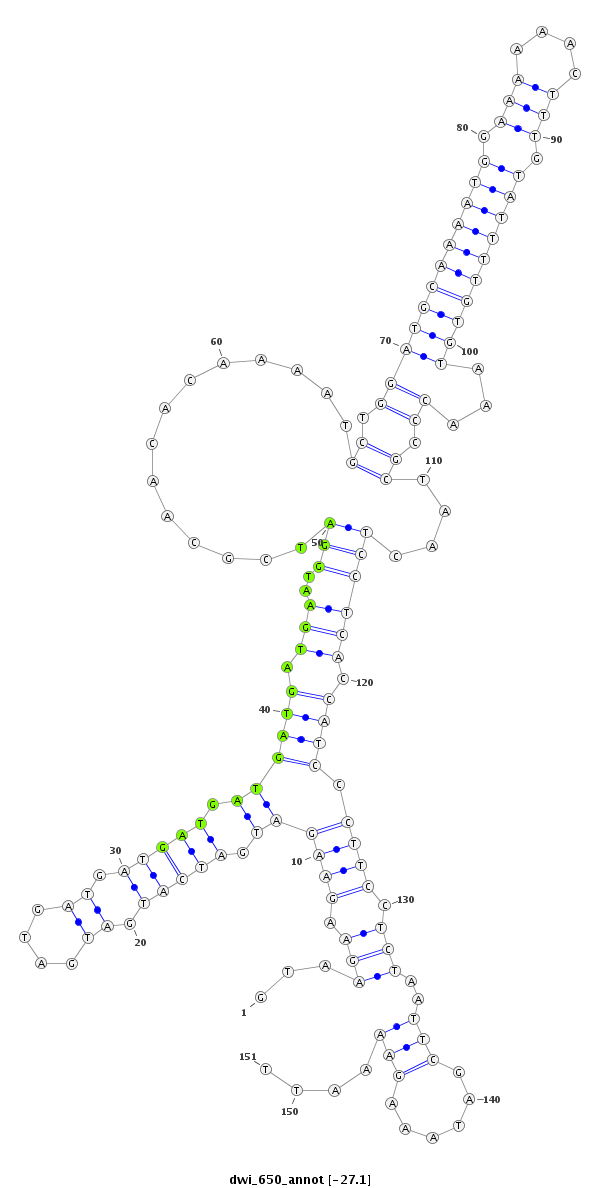
| ATGTGAACATCGAGTA-----CAT---------CCAGGACCTGAGCTCGCTCTCCCGCTTGGAGGTAA---------------------------------GGA-------------------------------------------------------------------------------------------------------------------------TCGTCCCGTTC--------CGTC-TCATCTCGGAATTCGAAT------TCGAAATTGGAAGCGGACCGGTATCGGTTAAGAAATCGGCATCGCATTTGGAGCCCGTT-TCACCAAATCCCC------AA-AGACGTTGGAATTCCGACCAGGGA--------------------ATAGGAATACGAAATGCGTGTGCGCCA----------------------AATT--TTCTTCAAAAT | Size | Hit Count | Total Norm | Total | M023
Head | M024
Male-body | M025
Embryo | M053
Female-body | O001
Testis | O002
Head | SRR553485
Ovary | SRR553486
Ovary | SRR553487
Ovary | SRR553488
Ovary | SRR618934
Ovary | V044
Embryo | SRR902008
Ovary | SRR902009
Testis | SRR1275487
Male larvae | SRR1275485
Male prepupae | SRR1275483
Male prepupae |
| ...................................AGGACCTGAGCTCGCTCTCCCGCT...................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 5.00 | 5 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................AGGACCTGAGCTCGCTCTCC.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................AT---------CCAGGACCTGAGCTCGCTCTCCCGC....................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......ACATCGAGTA-----CAT---------CCAGGACCTG...................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 3.00 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................AT---------CCAGGACCTGAGCTCGCTCTCCCGCT...................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 3.00 | 3 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CAGGACCTGAGCTCGCTCTCC.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 3.00 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........CGAGTA-----CAT---------CCAGGACCT....................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 3.00 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GAACATCGAGTA-----CAT---------CCAGGAC......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 3.00 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........CGAGTA-----CAT---------CCAGGACCTG...................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............A-----CAT---------CCAGGACCTGAGCTCGCT.............................................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................T---------CCAGGACCTGAGCTCGCTCT............................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........ATCGAGTA-----CAT---------CCAGGACCTG...................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................CAT---------CCAGGACCTGAGCTCGCTCTCCCGC....................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GAACATCGAGTA-----CAT---------CCAGGACCT....................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................CAT---------CCAGGACCTGAGCTCGCT.............................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CAGGACCTGAGCTCGCTCTCCC......................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CGCTCTCCCGCTTGGAGGAC.............................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 2 | 1.50 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GTA-----CAT---------CCAGGACCTGAGCT.................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................AT---------CCAGGACCTGAGCTCGCTCT............................................................................................................................................................................................................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................GAGCTCGCTCTCCCGCTTGGAG................................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........TCGAGTA-----CAT---------CCAGGACCTGAGCTCGCC.............................................................................................................................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................T---------CCAGGACCTGAGCTCGCTCTCCCGCT...................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................CCAGGACCTGAGCTCGCTCTCCC......................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................CAT---------CCAGGACCTGAGCTCGCTCTCC.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........CGAGTA-----CAT---------CCAGGACCTGAGCTCGCT.............................................................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CAGGACCTGAGCTCGCTCT............................................................................................................................................................................................................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................AT---------CCAGGACCTGAGCTCGCT.............................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................AGGACCTGAGCTCGCTCTCCC......................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TCGAGTA-----CAT---------CCAGGACCTGAGCTC................................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CAGGACCTGAGCTCGCTCTCCCGC....................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............AGTA-----CAT---------CCAGGACCTGAGCTCGCT.............................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......CATCGAGTA-----CAT---------CCAGGACCTG...................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............A-----CAT---------CCAGGACCTGAGCTC................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................GTTAAGAAATCGGCATCGCATTTGG............................................................................................................................................. | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........ATCGAGTA-----CAT---------CCAGGACCTGAGCTC................................................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TCGAGTA-----CAT---------CCAGGACCTG...................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................AGGACCTGAGCTCGCTCTCCCGCG...................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................GAAATTGGAAGCGGA............................................................................................................................................................................... | 15 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................AATACGAAATGCGTT.............................................. | 15 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |