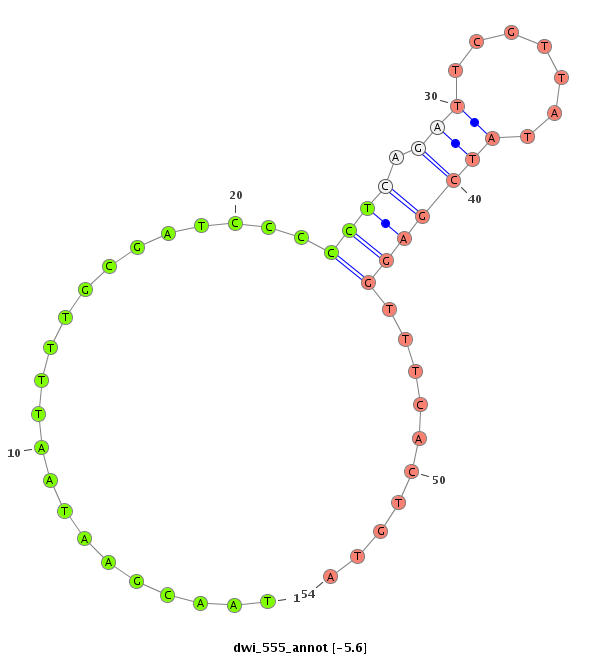
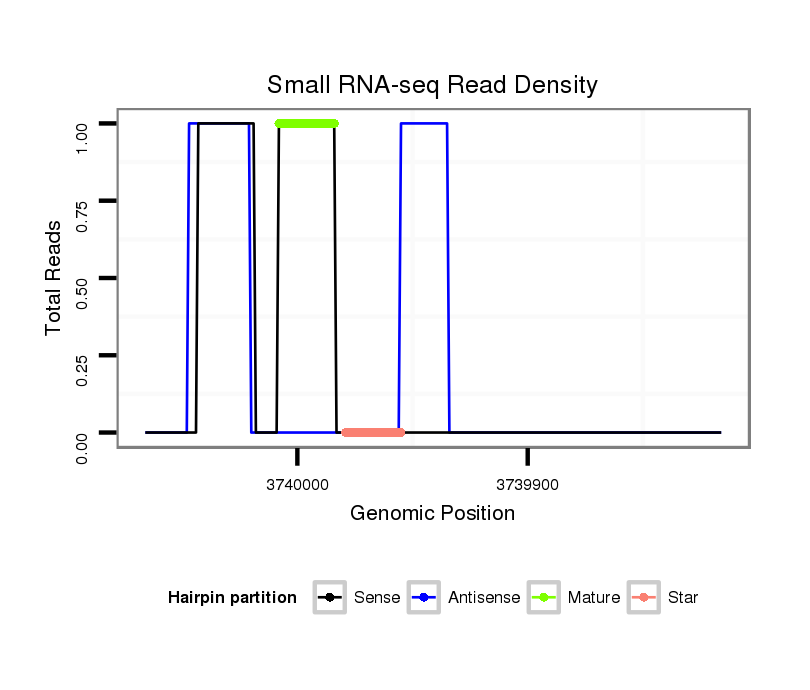
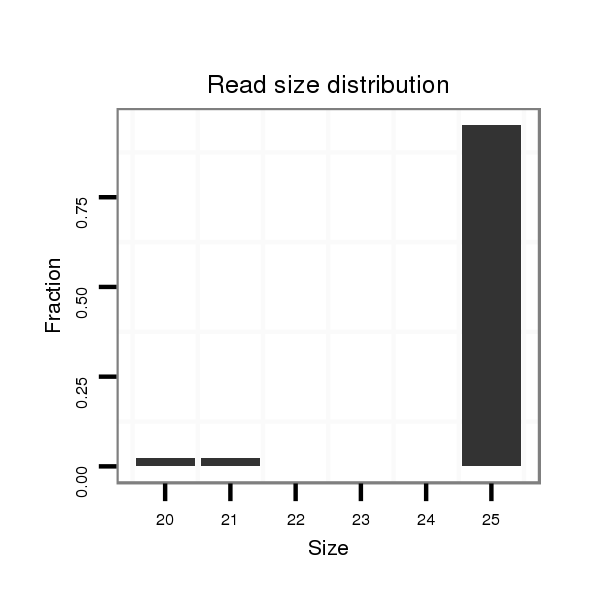
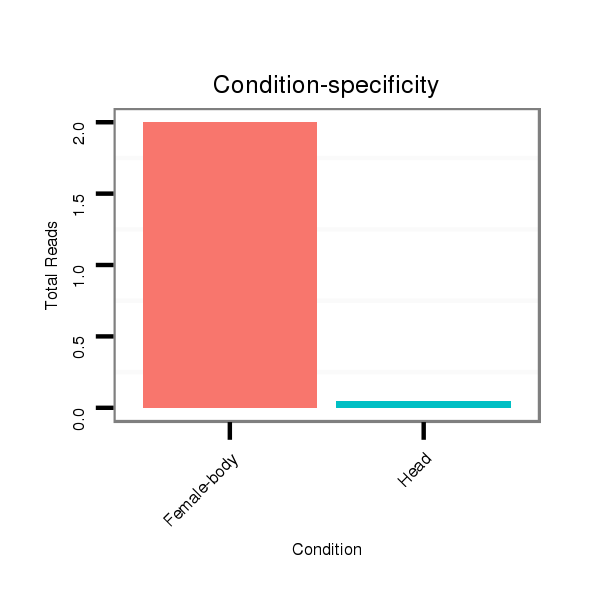
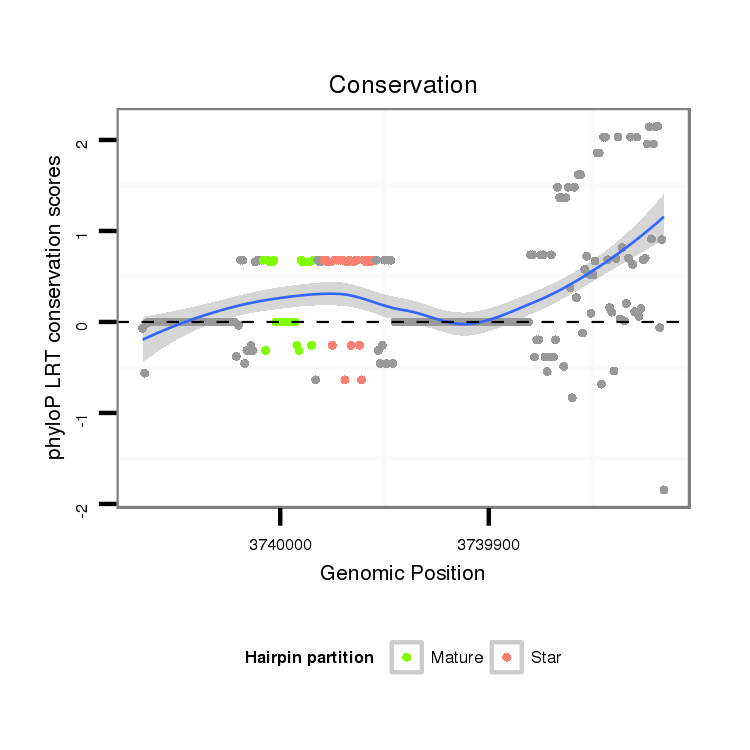
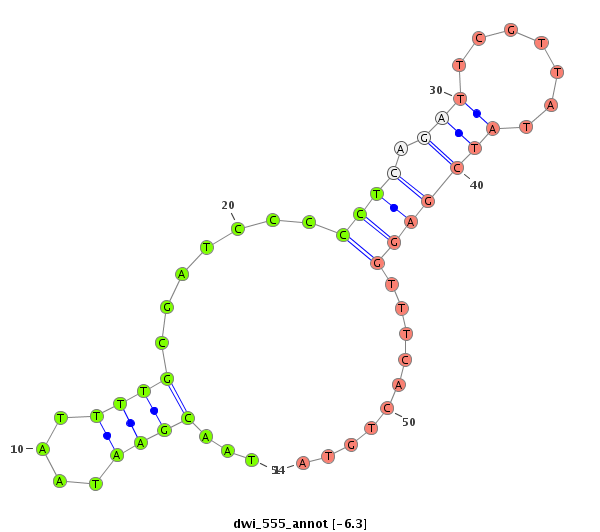
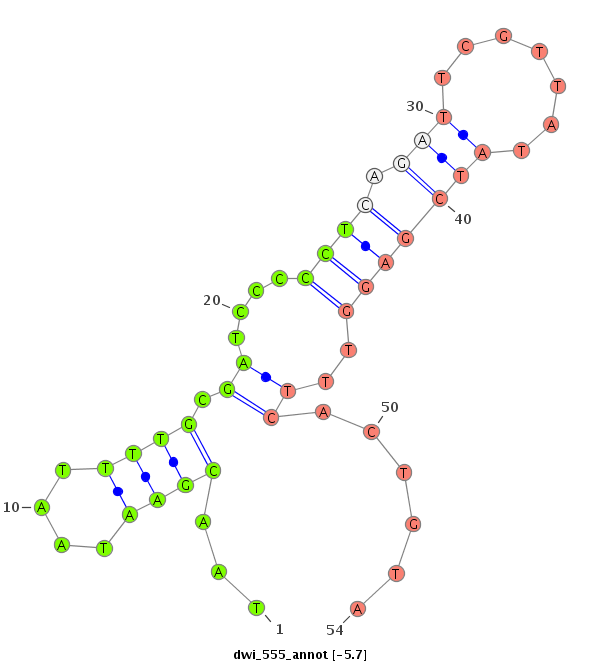
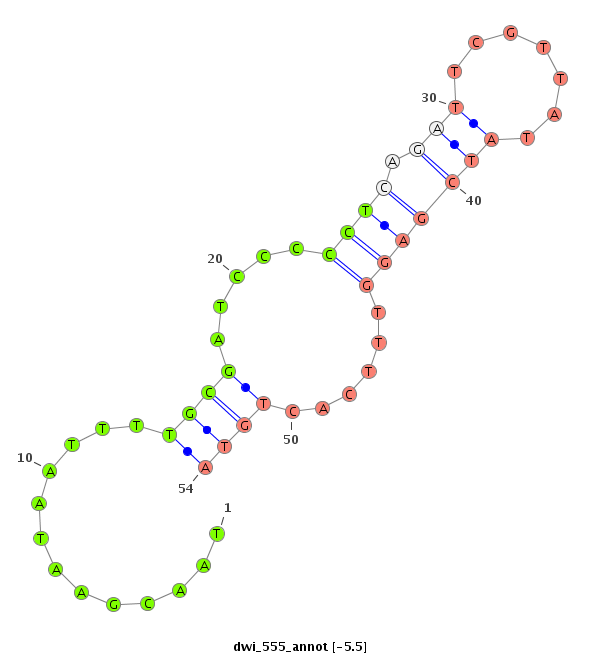
ID:dwi_555 |
Coordinate:scf2_1100000004511:3739866-3740016 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -6.3 | -5.7 | -5.5 |
 |
 |
 |
CDS [Dwil\GK16886-cds]; exon [dwil_GLEANR_17232:3]; intron [Dwil\GK16886-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AAGTCCCTTGAAAGGACTAAGGTTTTACATTTCAGTACTTATAGCGAATCAATAGATATAACGAATAATTTTGCGATCCCCCTCAGATTCGTTATATCGAGGTTTCACTGTATACCTAAACTTATGAACTGATACTTACCTACAAAATTGCAATCTATTTGTATATAATTTGTGTTTTTTTTCGTTTTTTTTTTGGTTTAGCACATTTTCCCGACAAGGGTTATTCCCTTGAACTGATAGAGCATGATATT **********************************************************......................((((.(((.......)))))))..........******************************************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
M020 head |
|---|---|---|---|---|---|---|---|
| ..........................................................TAACGAATAATTTTGCGATCCCCCT........................................................................................................................................................................ | 25 | 0 | 20 | 1.75 | 35 | 35 | 0 |
| .......................TTTACATTTCAGTACTTATAGCGAA........................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ...............................................................................................................................................................................................................TTCGCGACAATGGTTATTCC........................ | 20 | 2 | 4 | 0.25 | 1 | 0 | 1 |
| .......................................................................................TTCGTTATATCGAGGTTTCACTGTA........................................................................................................................................... | 25 | 0 | 20 | 0.20 | 4 | 4 | 0 |
| ............................................................................................................TGAATACGGAAACTTATGAAC.......................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 1 |
| .......................................TATAGCGAATCAATAGATATAACGA........................................................................................................................................................................................... | 25 | 0 | 20 | 0.15 | 3 | 3 | 0 |
| .............................................................................................ATATCGAGGTTTCACTGTAT.......................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| ...........................................................AACGAATAATTTTGCGATCCC........................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| ..................................................................AATTTTGCGATCCCCCTCAGATTCT................................................................................................................................................................ | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 |
| .......TTGAAAGGACTAAGGTTTTAA............................................................................................................................................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 |
| ............................................................................TCCCCCTCAGAGTCGTTATATCGAGG..................................................................................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 |
|
TTCAGGGAACTTTCCTGATTCCAAAATGTAAAGTCATGAATATCGCTTAGTTATCTATATTGCTTATTAAAACGCTAGGGGGAGTCTAAGCAATATAGCTCCAAAGTGACATATGGATTTGAATACTTGACTATGAATGGATGTTTTAACGTTAGATAAACATATATTAAACACAAAAAAAAGCAAAAAAAAAACCAAATCGTGTAAAAGGGCTGTTCCCAATAAGGGAACTTGACTATCTCGTACTATAA
*******************************************************************************************************************************************......................((((.(((.......)))))))..........********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V119 head |
V118 embryo |
V117 male body |
M020 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ...................TCCAAAATGTAAAGTCATGAATATCGC............................................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TATGGATTTGAATACTTGACT....................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................AACAAGGGAACTAGACTAT............ | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................ATAGCTCCAAAGTGACATAT......................................................................................................................................... | 20 | 0 | 20 | 0.15 | 3 | 0 | 1 | 2 | 0 | 0 |
| ....................................................................................................CCAAAGGGACGTGTGGATTT................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................GCAATATAGCTCCAAAGTGACAT........................................................................................................................................... | 23 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| .........................................................................GCTAGGGGGAGTCTAAGCA............................................................................................................................................................... | 19 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................AAAAGCGCTGTTACCAAT............................ | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................TCGCTTAGTTATCTATATTG............................................................................................................................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................AAAACGCTAGGGGGAGTCTAAGCA............................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................TAAGCAATATAGCTCCAAAGTGACAT........................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................CAATATAGCTCCAAAGTGACAT........................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................AAATAATGGAACTAGACTAT............ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................TATAGCTCCAAAGTGACATAT......................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................ATCGCTTAGTTATCTATATTGCTTAT........................................................................................................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................ATATAGCTCCAAAGTGACATAT......................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..CAGGGAACTTTCCTGATTCCAAAATGT.............................................................................................................................................................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................CAATATAGCTCCAAAGTGACATA.......................................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................GGGAGTCTAAGCAATATA.......................................................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................TTATTAAAACGCTAGGGGGAGTCTA................................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........TTTCCTGATTCCAAAATG............................................................................................................................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................CGAAAAAAAGCAAAAAAAAA.......................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................GGGGGAGTCTAAGCAATAT........................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................AAAAAAAAGCAAAAAAAA........................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004511:3739816-3740066 - | dwi_555 | AAGTCCCTTGAAAGGACTAAGGTTTTACATTTCAGTACTTATAGCGAATCAATAG----------ATATAACGAATAATTTTGCGATCCCCCTCAGATTCGTTATATCGAGGTTTCACTGTATACCTAAACTTATGAACTGATACTTACCTACAAAATTGCAATCTATTTGTATATAATTTGTGTTTTTTTTCGTTTTTTTTTTGGTTTAGCACATTTTCCCGACAAGGGTTATTCCCTTGAACTGATAGAGCATGATATT |
| droVir3 | scaffold_13049:8326650-8326658 + | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATGATATT | |
| droMoj3 | scaffold_6680:11980628-11980669 + | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCTTATAATGGCTACAAATTGAATTTGATCAGACATGATATA | |
| droGri2 | scaffold_15110:7354907-7354945 + | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATAATGGTTACAAGCTGAACCTGATGCGGCATGATATA | |
| dp5 | 4_group3:3300791-3300873 + | ATG------------------------------------------GTATATTCTGATAAAATTATATATTACGA----------ATTCCCCTTGAGATTCGCTATATGGAAGTTCGACTGTATTATTCAAA------------------------------------------------------------------------------------------------------------------------------TTGT | |
| droPer2 | scaffold_61:50822-50873 - | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCCCAACTGCCCGGCCTTGGGTATCGCCTGAGTCTGATCGAGCACGATATC | |
| droAna3 | scaffold_13337:12765099-12765150 - | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCTCAACTGCCGGACTTTGGCTATCGCCTCACTCTCATCGAGCATGATGTA | |
| droBip1 | scf7180000396589:1257907-1257958 + | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCTCAACTGCCGGACTTTGGGTATCGCCTCACTCTCGTCGAGCATGATGTT | |
| droKik1 | scf7180000302686:73587-73638 + | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCTCAAGTGCCGGACTTTGGCTATCGGCTCAGTCTCATCGAGCATGATATA | |
| droFic1 | scf7180000453929:1840448-1840517 - | TCA------------------------------------------CT-----------------------------------------------------------------------------------------------------------------------------------------------------TTGCCTTGCTTGCAGCCCAAATGCCGGACTTTGGCTATCGGCTCAGTCTCGTCGAGCATGATGTA | |
| droEle1 | scf7180000491268:1273119-1273151 + | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCTATCGGCTCAGTCTCGTCGAGCATGATATA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
Generated: 05/17/2015 at 11:29 PM