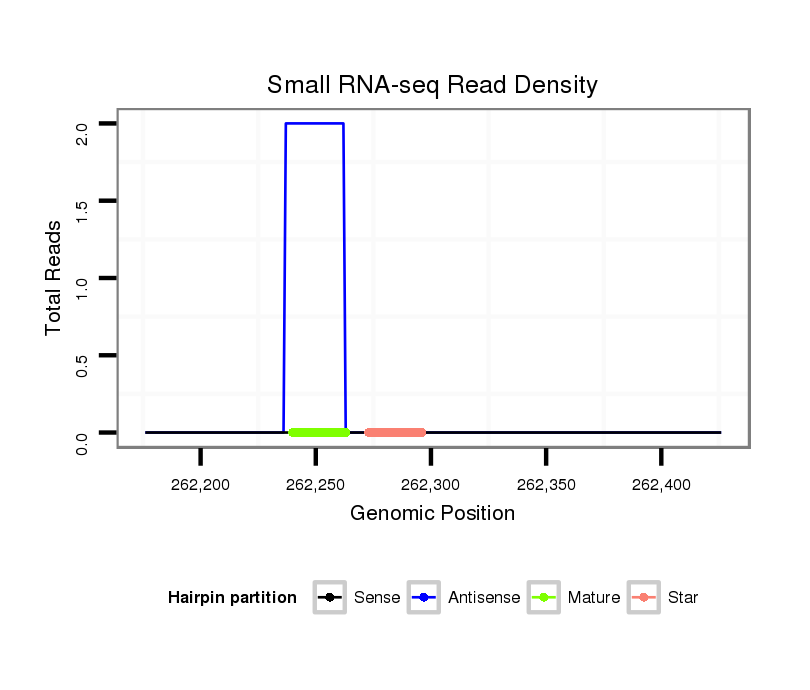
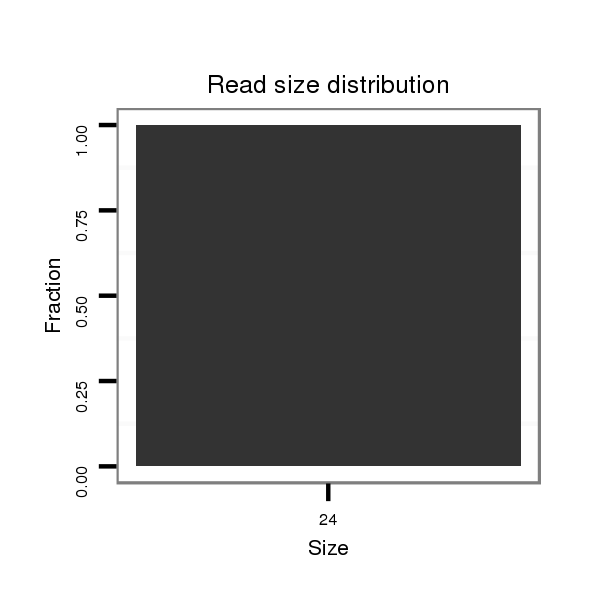
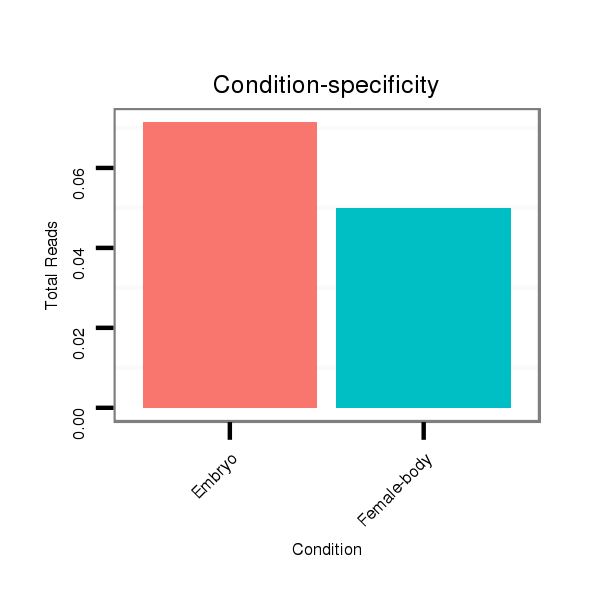
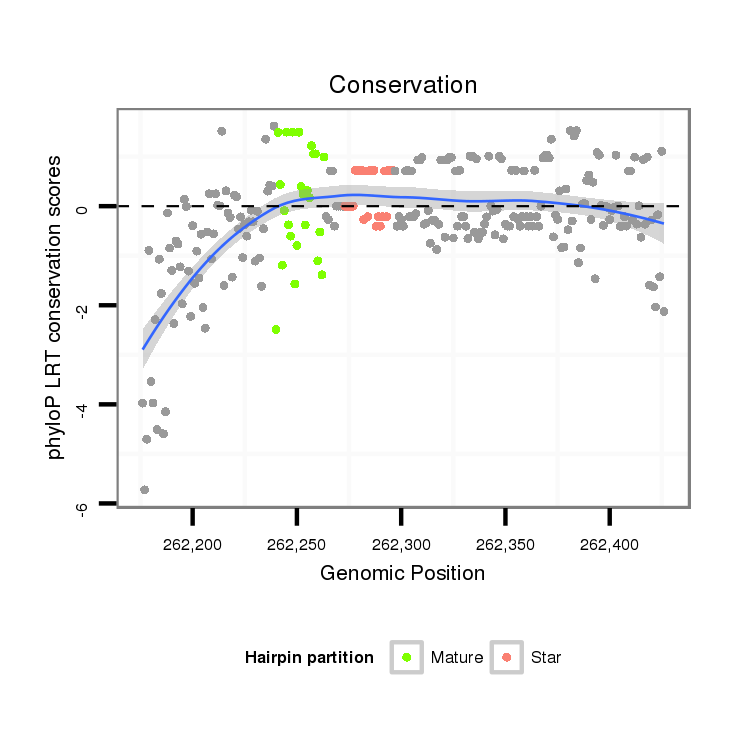
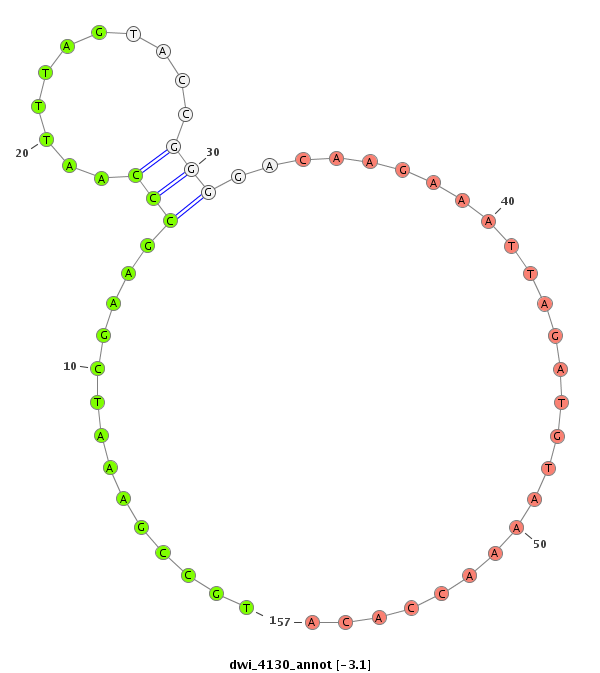
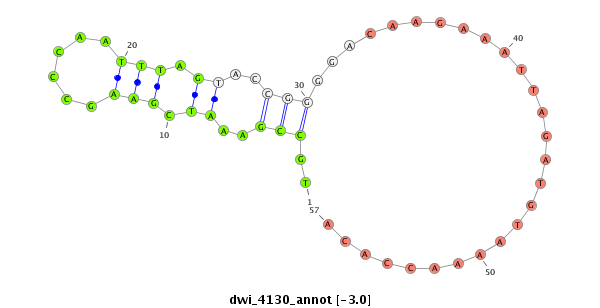
| .............................................................TGAACGGCTTTAGCTTCGGGTTAAAT.................................................................................................................................................................... |
26 |
0 |
14 |
2.14 |
30 |
23 |
4 |
2 |
1 |
| ..............................................................GAACGGCTTTAGCTTCGGGTTAAAT.................................................................................................................................................................... |
25 |
0 |
14 |
0.86 |
12 |
9 |
3 |
0 |
0 |
| ..................TCAAGCGGAGTTCCCTCCTAGTAGT................................................................................................................................................................................................................ |
25 |
0 |
20 |
0.70 |
14 |
0 |
14 |
0 |
0 |
| ..............GCGCTCAAGCGGAGTTCCCTCCTAGT................................................................................................................................................................................................................... |
26 |
0 |
19 |
0.53 |
10 |
1 |
9 |
0 |
0 |
| .......................................................TGCAGCTGAACGGCTTTAGCTTCGGGT......................................................................................................................................................................... |
27 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
| ........................................................TCAGCTGAACGGCTTTAGCTTCGGGT......................................................................................................................................................................... |
26 |
0 |
13 |
0.46 |
6 |
4 |
2 |
0 |
0 |
| .............GGCGCTCAAGCGGAGTTCCCTCCTAGT................................................................................................................................................................................................................... |
27 |
0 |
19 |
0.26 |
5 |
3 |
2 |
0 |
0 |
| ................GCTCAAGCGGAGTTCCCTCCTAGTAGT................................................................................................................................................................................................................ |
27 |
0 |
20 |
0.25 |
5 |
1 |
4 |
0 |
0 |
| .......................................................TTCAGCTGAACGGCTTTAGCTTCTGGT......................................................................................................................................................................... |
27 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
| ............................................................CTGAACGGCTTTAGCTTCGGGTTAAAT.................................................................................................................................................................... |
27 |
0 |
14 |
0.21 |
3 |
3 |
0 |
0 |
0 |
| ...............CGCTCAAGCGGAGTTCCCTCCTAGT................................................................................................................................................................................................................... |
25 |
0 |
19 |
0.16 |
3 |
0 |
3 |
0 |
0 |
| .....TTAGCAAAGGCGCTCAAGCGGAGT.............................................................................................................................................................................................................................. |
24 |
0 |
20 |
0.15 |
3 |
3 |
0 |
0 |
0 |
| ........................CGAGTTCCCTCCTAGTAGTAGGACT.......................................................................................................................................................................................................... |
25 |
1 |
20 |
0.15 |
3 |
0 |
3 |
0 |
0 |
| ................GCTCAAGCGGAGTTCCCTCCTAGT................................................................................................................................................................................................................... |
24 |
0 |
20 |
0.15 |
3 |
0 |
3 |
0 |
0 |
| .................CTCAAGCGGAGTTCCCTCCTAGTAGT................................................................................................................................................................................................................ |
26 |
0 |
20 |
0.15 |
3 |
1 |
2 |
0 |
0 |
| ..................TCAAGCGGAGTTCCCTCCTAGTAG................................................................................................................................................................................................................. |
24 |
0 |
20 |
0.15 |
3 |
0 |
3 |
0 |
0 |
| ..............................................................GAACGGCTTTAGCTTCTGGTTAAATTTT................................................................................................................................................................. |
28 |
3 |
9 |
0.11 |
1 |
0 |
1 |
0 |
0 |
| ...............CGCTCAAGCGGAGTTCCCTCCTAGTAGT................................................................................................................................................................................................................ |
28 |
0 |
19 |
0.11 |
2 |
0 |
2 |
0 |
0 |
| ........................................................................................................AATCTACATTTTGGTGTCAGATCTCGT........................................................................................................................ |
27 |
0 |
20 |
0.10 |
2 |
0 |
2 |
0 |
0 |
| ..TCCTTAGCAAAGGCGCTCAAGCGGAGTT............................................................................................................................................................................................................................. |
28 |
0 |
10 |
0.10 |
1 |
0 |
1 |
0 |
0 |
| .........CAAAGGCGCTCAAGCGGAGT.............................................................................................................................................................................................................................. |
20 |
0 |
20 |
0.10 |
2 |
0 |
2 |
0 |
0 |
| .......AGCAAAGGCGCTCAAGCGGAGT.............................................................................................................................................................................................................................. |
22 |
0 |
20 |
0.10 |
2 |
1 |
1 |
0 |
0 |
| ......................GCGGAGTTCCCTCCTAGTAGTAGGACT.......................................................................................................................................................................................................... |
27 |
0 |
20 |
0.10 |
2 |
0 |
2 |
0 |
0 |
| .............................................................TGAACGGCTTTAGCTTCGGGTTAAATC................................................................................................................................................................... |
27 |
0 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
| .........................................................CAGCTGAACGGCTTTAGCTTCGGGT......................................................................................................................................................................... |
25 |
0 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
| ........................................................TCAGCTGAACGGCTTTAGCTTCGGGTT........................................................................................................................................................................ |
27 |
0 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
| .............................................................TGAACGGCTTTAGCTTCGGGT......................................................................................................................................................................... |
21 |
0 |
14 |
0.07 |
1 |
1 |
0 |
0 |
0 |
| .............................................................TGAACGGTTTTAGCTTCGGGTTAAAT.................................................................................................................................................................... |
26 |
1 |
17 |
0.06 |
1 |
1 |
0 |
0 |
0 |
| ..........AAAGGCGCTCAAGCGGAGTTCCCTCCT...................................................................................................................................................................................................................... |
27 |
0 |
19 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ............AGGCGCTCAAGCGGAGTTCCCTCCTAGT................................................................................................................................................................................................................... |
28 |
0 |
19 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ..........CAAGGGGCCCAAGCGGAGTT............................................................................................................................................................................................................................. |
20 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
| .................CTCAAGCCGAGTTCCCTCCTAGTAGT................................................................................................................................................................................................................ |
26 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................GGCCCCTGTTCTTTAATCTACAT.......................................................................................................................................... |
23 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| .......................................................................................................TAATCTACATTTTGGTGTCAGATCTCGT........................................................................................................................ |
28 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| .................AACAAGCCGAGTTCCCTCCTAGTAGT................................................................................................................................................................................................................ |
26 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ........................CGACTTCCCTCCTAGTAGTAGGACT.......................................................................................................................................................................................................... |
25 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ...............................CCTCCTAGTAGTAGGACTC......................................................................................................................................................................................................... |
19 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ...........................GTTCCCTCCTAGTAGTAGGACTCCA....................................................................................................................................................................................................... |
25 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ............AGGTGCCCAAGCGGAGTG............................................................................................................................................................................................................................. |
18 |
3 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
| .................................................................CGGCTTTAGCTTCGGGTTAA...................................................................................................................................................................... |
20 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| .................CTCAAGCGGAGTTCCCTCCTAGTAG................................................................................................................................................................................................................. |
25 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| .....................TGCGGAGTTCCCTCCTAGTAGTAGGACT.......................................................................................................................................................................................................... |
28 |
1 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
| .................................................................................................................................................................GCTGGTGGCCATGGTGTTT....................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ...............TGCTCAAGCGGAGTTCCCTCCTAGTAGT................................................................................................................................................................................................................ |
28 |
1 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ..................TCAAGCCGAGTTCCCTCCTAGTAGC................................................................................................................................................................................................................ |
25 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| .........................................................................................................ATCTACATTTTGGTGTCAGATCT........................................................................................................................... |
23 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ................GTTCAAGCGGAGTTCCCTCCTAGTAGT................................................................................................................................................................................................................ |
27 |
1 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ..................ACAAGCCGAGTTCCCTCCTAGTAGT................................................................................................................................................................................................................ |
25 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| .....................AGCGGAGTTCCCTCCTAGTAGTAGGACT.......................................................................................................................................................................................................... |
28 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ......TAGCAAAGGCGCTCAAGCGGAGTTCCCT......................................................................................................................................................................................................................... |
28 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ......................TCGGAGTTCCCTCCTAGTAGTAGGACT.......................................................................................................................................................................................................... |
27 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ..................TCAACCGGAGTTCCCTCCTAGTAGT................................................................................................................................................................................................................ |
25 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |