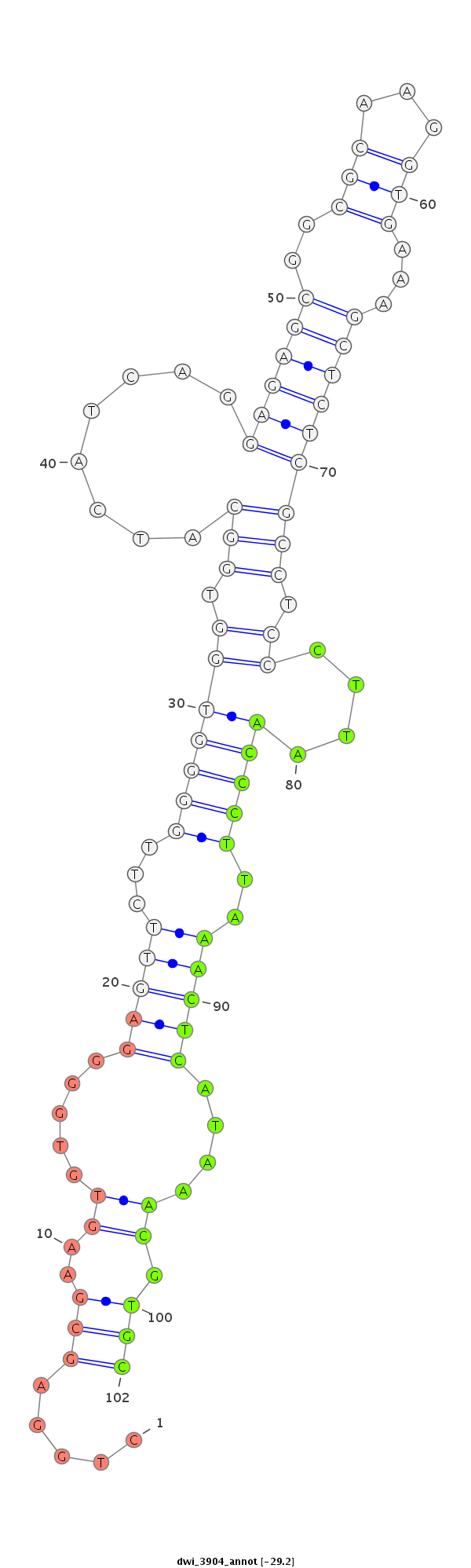
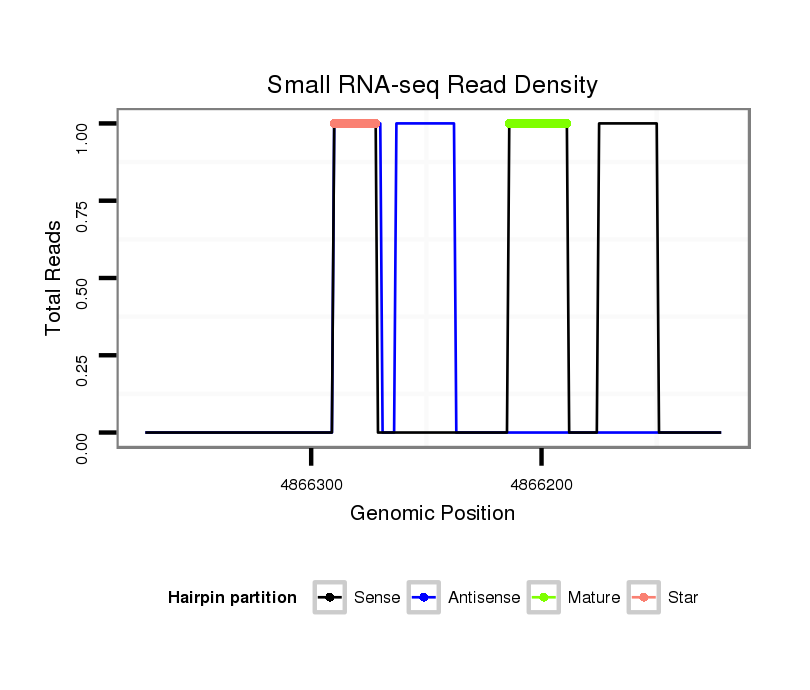
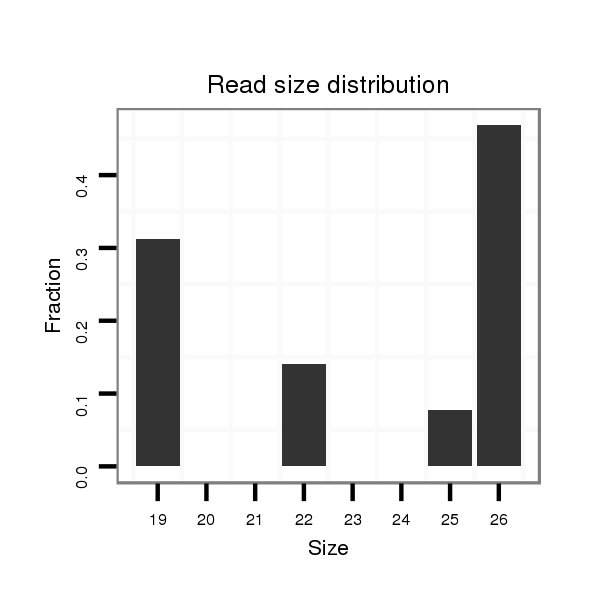
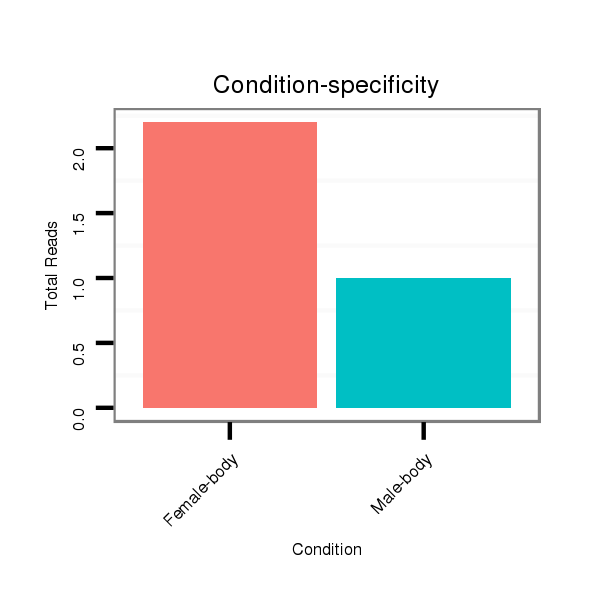
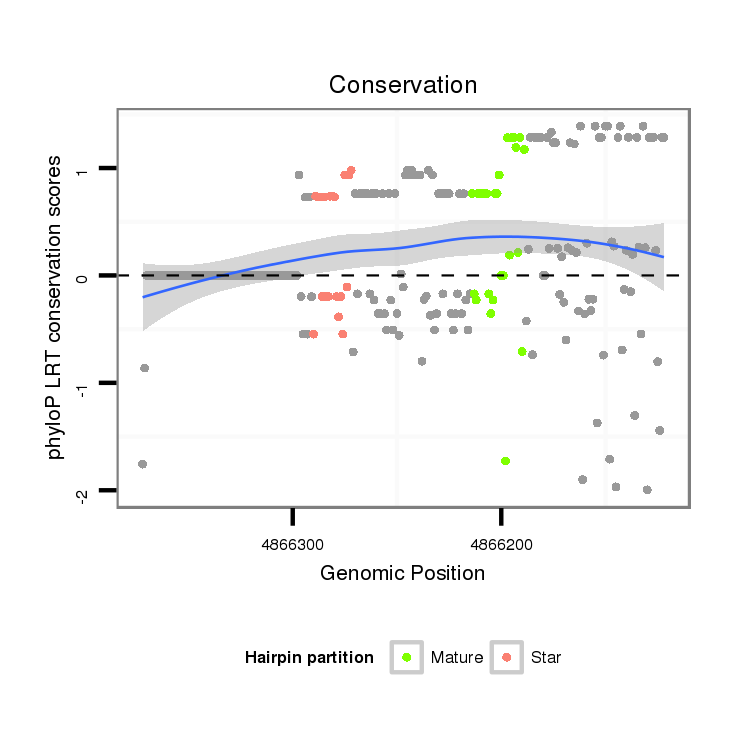
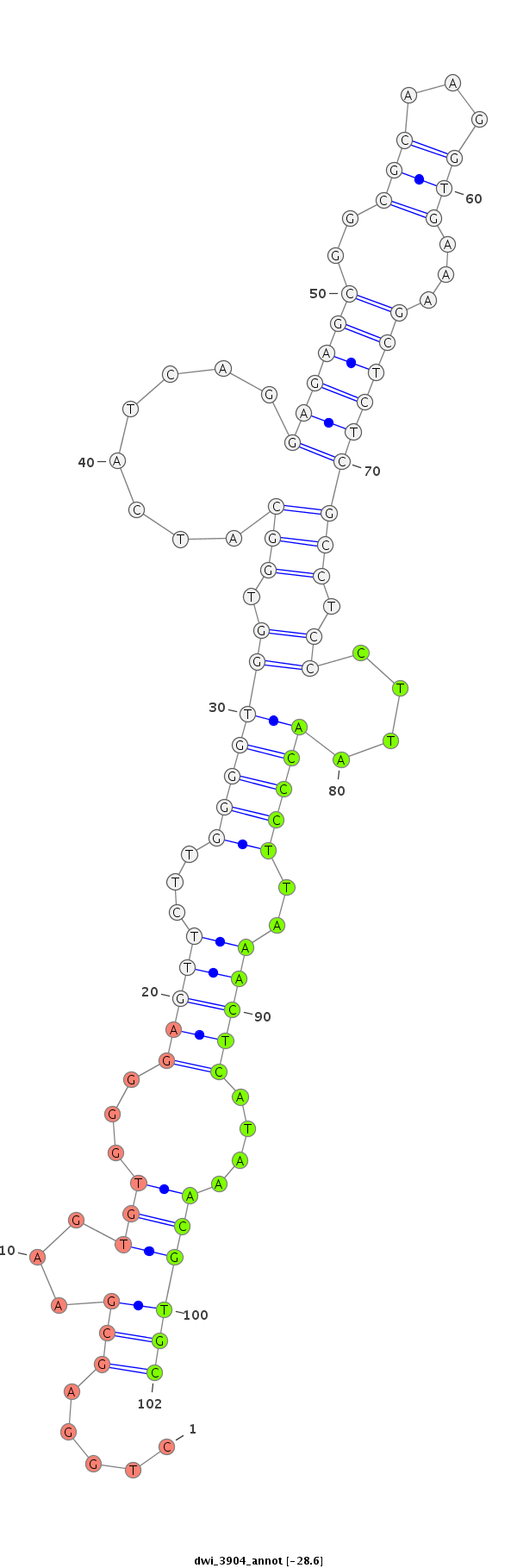
ID:dwi_3904 |
Coordinate:scf2_1100000004961:4866172-4866322 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -28.6 | -28.5 | -28.3 |
 |
 |
 |
CDS [Dwil\GK23360-cds]; exon [dwil_GLEANR_6705:3]; intron [Dwil\GK23360-in]
| Name | Class | Family | Strand |
| BEL-10_DWil-I-int | LTR | Pao | + |
| BEL-10_DWil-LTR | LTR | Pao | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GGAAATCTCTCTTCACTTTTTCGTTTATTCATTTTTCTAGTTGAACTAAATCTAGCTTATTTAAGAGCTTAATAAGCCCCGCCTGGAGCGAAGTGTGGGGAGTTCTTGGGGTGGTGGCATCATCAGGAGAGCGGCGCAAGGTGAAAGCTCTCGCCTCCCTTAACCCTTAAACTCATAAACGTGCTCCACCCCCCCTTGCAGGAGTGCAAGACCAGGAAGCCGAAGACTGTGCGCATCGCGATTGGCCGACC **********************************************************************************.....(((..((.....(((((...(((((((.(((........((((((..(((...)))...))))))))).))....)))))..)))))....)).)))******************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
V118 embryo |
V119 head |
|---|---|---|---|---|---|---|---|---|---|
| ................................................................................CGCTGGAGCGAAGTGTGGGGAGTTCTT................................................................................................................................................ | 27 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................GCAGGAGTGCAAGACCAGGAAGCCGA............................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................CTTAACCCTTAAACTCATAAACGTGC................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................AGACTGTGCGCATCGCGATTGGCCGT.. | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..................................................................................CTGGAGCGAAGTGTGGGGA...................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................CAAGGTGAAAGCCATCGCCTGC............................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................GGTGGGGAGTTTCTGGGGTGG......................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................TAATAAGCGGCGCCTGGAGCA................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................TGAAAGCTCTCGCCTCCCTTAACCCT.................................................................................... | 26 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................TGGAGCGAAGTGTGGGGAGTTC.................................................................................................................................................. | 22 | 0 | 20 | 0.45 | 9 | 9 | 0 | 0 | 0 |
| ..............................................................................................GTGGGGAGTTTCTGGGGCGG......................................................................................................................................... | 20 | 3 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 |
| ................................................................................................GGGGAGTTCCTGGGGCGG......................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................GGAGAGCGGCGCAAGGTGAAAGCTC..................................................................................................... | 25 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................GTGGGGAGTTGCTGGGGCGG......................................................................................................................................... | 20 | 3 | 16 | 0.25 | 4 | 2 | 2 | 0 | 0 |
| ..................................................................................CTGGAGCAAAGTGTGGGG....................................................................................................................................................... | 18 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................TGGAGCAAAGTGTGGGGAGTTCTTG............................................................................................................................................... | 25 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................GGTGGGGAGTTTTTGGGGCGG......................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TGCAGGAGTGCAAGACCAGGA.................................. | 21 | 0 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................TTGCAGGAGTGCAAGACCAGGA.................................. | 22 | 0 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 |
| ..............................................................................CCGCCGGTAGCTAAGTGTG.......................................................................................................................................................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................GAAAGGTCTCGCCTTCCTG.......................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................CCTGGGGTGAAGTGTGGGGG...................................................................................................................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................TAGAGCGAAGTGTGGGGAGTTCT................................................................................................................................................. | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................TGGAGCGAAGTGTGGGGACG.................................................................................................................................................... | 20 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................ACAGGGGCGCATGGTGAA.......................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
CCTTTAGAGAGAAGTGAAAAAGCAAATAAGTAAAAAGATCAACTTGATTTAGATCGAATAAATTCTCGAATTATTCGGGGCGGACCTCGCTTCACACCCCTCAAGAACCCCACCACCGTAGTAGTCCTCTCGCCGCGTTCCACTTTCGAGAGCGGAGGGAATTGGGAATTTGAGTATTTGCACGAGGTGGGGGGGAACGTCCTCACGTTCTGGTCCTTCGGCTTCTGACACGCGTAGCGCTAACCGGCTGG
*******************************************************************.....(((..((.....(((((...(((((((.(((........((((((..(((...)))...))))))))).))....)))))..)))))....)).)))********************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
V118 embryo |
|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................TCCTCACGTTGTGCTCCTG................................. | 19 | 3 | 20 | 1.15 | 23 | 22 | 1 | 0 |
| .............................................................................................................CCACCACCGTAGTAGTCCTCTCGCCG.................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................................................................GACCTCGCTTCACACCCCTCA.................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................TCCTCACGTTGTGCTCCT.................................. | 18 | 2 | 9 | 0.33 | 3 | 3 | 0 | 0 |
| ...................................................................................................................................................................................GCACGAGGTGGGGCGCAAA..................................................... | 19 | 3 | 6 | 0.33 | 2 | 1 | 1 | 0 |
| ...................................................................................................................................................................................................................................AGACGCGTAGTGCTAACCGC.... | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 |
| ....................................................................................................................................................AAAGCGGAGGGAATTGGGAA................................................................................... | 20 | 1 | 8 | 0.13 | 1 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................CCTCACGTTGTGCTCCTG................................. | 18 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| .........................................................................................................................................TTCCACTCGCGAGAGCAGA............................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 |
| ........................................................................TTTCGGGGCCGACCGCGC................................................................................................................................................................. | 18 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 |
| .............................................................................................................................................................................................GGGGGGAACGTCCTCACGTTC......................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ........................................................................................GCTTCACACCCCTCAAGAA................................................................................................................................................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ............................................................................................................................................................................................GGGGGGGAACGTCCTCACGTTCTG....................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ...................................................................................ACCTCGCTTCACACCCCT...................................................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004961:4866122-4866372 - | dwi_3904 | GGAAATCTCTCTTCACTTTTTCGTTTATTCATTTTTCTAGTTGAACTAAATCTAGCTTATTTAAGAGCTTAATAAGCCCCGCCTGGAGCGAAGTGTGGGGAGTTCTTGGGGT----GGTGGCATCATCAGGAGAGCG---GCGCAAGGTGAAAGCTCTCGCCTCCCTTAACCCTTAAACTCATAAACGTGCTCCACCCCCCCTTGCAG-------GAGTGC---A----------------------------AGACCAGGAAGCCGAAGACTGTGCGCATCGCGATTGGCCGACC---------- |
| dp5 | Unknown_group_6:40258-40351 - | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGAACATCCTCCACCCC--CTTGCAG-------GATTGC---A--------AGGCTGAGTGCACGCCCCCGGCAGGATCCATCCAAACACGGTGCTCTGAGCCACCGGCAGGCC---------- | |
| droPer2 | scaffold_43:209119-209212 - | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGAACATCCTCCACCCC--CTTGCAG-------GATTGC---A--------AGGTTGAGTGCACGCCCCCGGCAGGATCCATCCAAACACGGTGCTCTGAGCCACCGGCAGGCC---------- | |
| droFic1 | scf7180000453500:1301-1455 - | GA-------------------------------------------------------------------------GTGCGGTGTGGGGTAAAGCTCCGGGAG-------------------------AGGGAGAGCG---GGAGAGG-------------------------------C--CTGAACATT--CCGCCCC--CTTGCAACCGCTGGCGTTGC---AAATAACTAGCCCTGGTTGCAGGCGCCCGACAGGATCCAGCCGAAGATGGTCTTCTGAGCTACTGGCAATCC---------- | |
| droRho1 | scf7180000768343:5402-5499 + | C--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCCCCCCCCC--CCTGCGG-------GAGAGTTCC-TTGAAGCAGGACTGACGGCAGGCTCCCGACACGATCCATCCAAACGCCGTGCTCTGAGCGACGGGCAGCCC---------- | |
| droSec2 | scaffold_1949:3883-3954 - | C------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGAACATTCTCCCCCCC--C------------------------------------------CCGCTCCAGAAACGGTCCATCCGAACACCGTGTTCAGCGCGACGGGCAACCC---------- | |
| droYak3 | v2_chrUn_003:300769-300944 - | GC-------------------------------------------------------------------------G---------------------GAGACTCCTTGGAGACTCTGTGGTGAAGAGCGAGAGAGCGTAAGAGGAAGCGGAGAGCGGGTGACAGTCCAAACCCCGTAAC--TTGAACATTCCGC-CCCC--CTTGCAATCACTGGGGTTGC---A----------------------------ACAACAAAAACCCAAAAACAGTTTTCTGGGTGACCGGCATTCCCTCTTCGAAG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/19/2015 at 10:34 AM