
ID:dwi_3875 |
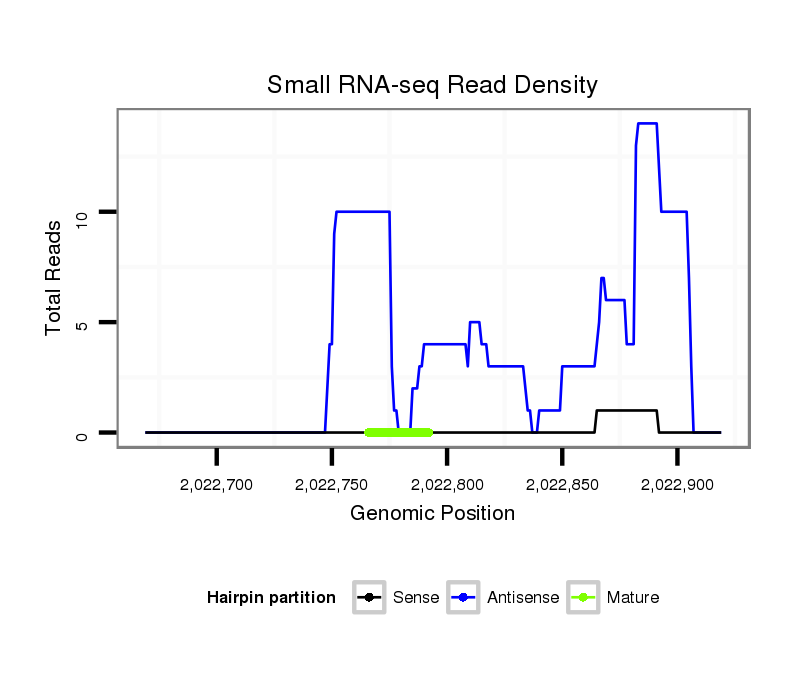
Coordinate:scf2_1100000004961:2022719-2022869 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -16.6 | -16.3 | -16.3 | -16.2 |
 |
 |
 |
 |
CDS [Dwil\GK23438-cds]; exon [dwil_GLEANR_6915:2]; intron [Dwil\GK23438-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTCGACCCCGGGCTAAAAGATAATTTTTTTTTTTATTTTGTAATAATTTAATTTTTTTTCTCACAAAAAAAAAACATTAGCAACTAGTTTAAAAATAATTGACTTCAAGAACAGCATTACGTCGGAAATTCATAAGAGCTATGTAAGTACACAGCTGTTGTATGAAAAGCACAAAATTGTAATTCTTTTTTATATTTGCAGGCCTCTCAAGTAGCTGGAGACAACTCCCGCTTGTGAAGAGCCACAACCTG **************************************************.........((..................(((...(((.(((((............(((((..(((............(((((((.((((.(((....))).)))).)))..))))...........)))..)))))))))).))))))))************************************************** |
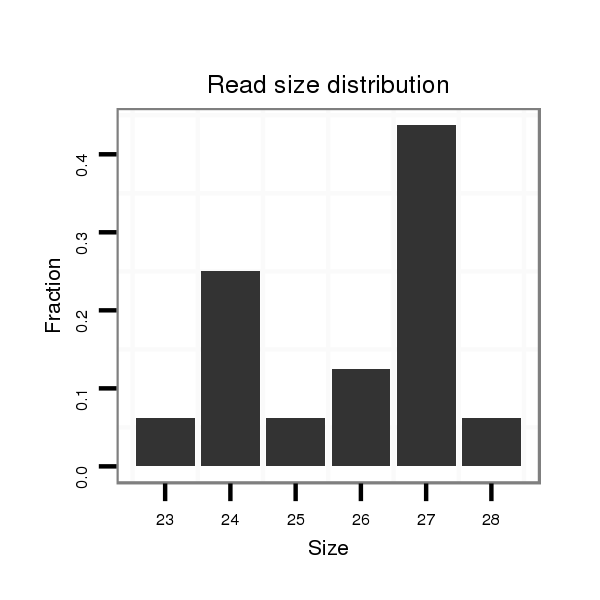
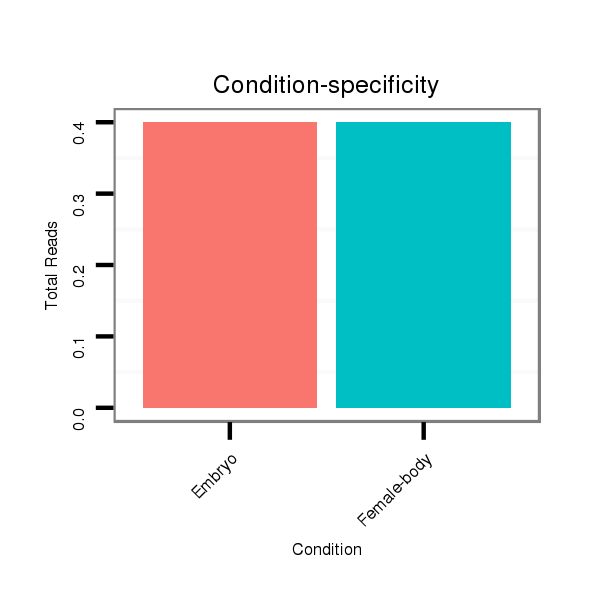
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
M045 female body |
|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................TGCAGGCCTCTCAAGTAGCTGGAGACA............................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ...........................................................................................................................................................................................................................GACAACTCCCGCTTGTGAAGAGCCAT...... | 26 | 1 | 2 | 0.50 | 1 | 0 | 1 |
| .................................................................................................ATTGACTTCAAGAACAGCATTACGTCG............................................................................................................................... | 27 | 0 | 20 | 0.35 | 7 | 3 | 4 |
| .................................................................................................ATTGACTTCAAGAACAGCATTACG.................................................................................................................................. | 24 | 0 | 20 | 0.20 | 4 | 3 | 1 |
| ...GACCGTGGCCTAAAAGATAA.................................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 |
| ..................................................................................................TTGACTTCAAGAACAGCATTACGTCG............................................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 |
| .................................................................................................ATTGACTTCAAGAACAGCATTACGTCGG.............................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| ..................................................................................................TTGACTTCAAGAACAGCATTACG.................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| .................................................................................................ATTGACTTCAAGAACAGCATTACGT................................................................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 |
|
AAGCTGGGGCCCGATTTTCTATTAAAAAAAAAAATAAAACATTATTAAATTAAAAAAAAGAGTGTTTTTTTTTTGTAATCGTTGATCAAATTTTTATTAACTGAAGTTCTTGTCGTAATGCAGCCTTTAAGTATTCTCGATACATTCATGTGTCGACAACATACTTTTCGTGTTTTAACATTAAGAAAAAATATAAACGTCCGGAGAGTTCATCGACCTCTGTTGAGGGCGAACACTTCTCGGTGTTGGAC
**************************************************.........((..................(((...(((.(((((............(((((..(((............(((((((.((((.(((....))).)))).)))..))))...........)))..)))))))))).))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V118 embryo |
V119 head |
M020 head |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................TGATCAAATTTTTATTAACTGAAGT................................................................................................................................................ | 25 | 0 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................CGACCTCTGTTGAGGGCGAACACTT............. | 25 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 |
| .....................................................................................................................................................................................................................CGACCTCTGTTGAGGGCGAACACT.............. | 24 | 0 | 1 | 3.00 | 3 | 0 | 2 | 1 | 0 |
| .....................................................................................................................................................................................................................CGACCTCTGTTGAGGGCGAACAC............... | 23 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 |
| .....................................................................................................................................................................................TAAGAAAAAATATAAACGTCCGGAGAGT.......................................... | 28 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...............................................................................CGTTGATCAAATTTTTATTAACTGAAGT................................................................................................................................................ | 28 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .............................................................................................................................................ACATTCATGTGTCGACAACATACT...................................................................................... | 24 | 0 | 2 | 1.50 | 3 | 2 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................GTCCGGAGAGTTCATCGACCTCTGT............................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................GTTGATCAAATTTTTATTAACTGAAGT................................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................ACATTCATGTGTCGACAACATACTTTT................................................................................... | 27 | 0 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 |
| .........................................................................................................................AGCCTTTAAGTATTCTCGATACATTCAT...................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GTTTTAACATTAAGAAAAAATATAAACGT................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................AATGCAGCCTTTAAGTATTCTCGAT.............................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................TGATCAAATTTTTATTAACTGAAGTTCT............................................................................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................GACCTCTGTTGAGGGCGAACACT.............. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................GTTGATCAAATTTTTATTAACTGAAGTT............................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................ACATTCATGTGTCGACAACATACTT..................................................................................... | 25 | 0 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 |
| .......................................................................................................................GCAGCCTTTAAGTATTCTCGATACATT......................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................ACGTCCGGAGAGTTCATCGACCTCTGTT........................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................CGTCCGGAGAGTTCATCGACCTCTGT............................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................GATCAAATTTTTATTAACTGAAGTT............................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................GTCCGGAGAGTTCATCGACCTCTGTT........................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................AATGCAGCCTTTAAGTATTCTCGA............................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................TCTTGTCGTAATGCAGCCTTTAAGTAT..................................................................................................................... | 27 | 0 | 19 | 0.53 | 10 | 6 | 3 | 0 | 1 |
| .............................................................................................................................TTTAAGTATTCTCGATACATTCATGT.................................................................................................... | 26 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................ACATTCATGTGTCGACAACATAC....................................................................................... | 23 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TCATCGACCTCTGTTGAGGGCGAAG................. | 25 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................TCATGTGTCGACAACATACTTTTCGT................................................................................ | 26 | 0 | 20 | 0.50 | 10 | 8 | 2 | 0 | 0 |
| ...........................................................................................................................................ATACATTCATGTGTCGACAACATACTT..................................................................................... | 27 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................CTTGTCGTAATGCAGCCTTTAAGTAT..................................................................................................................... | 26 | 0 | 19 | 0.32 | 6 | 4 | 2 | 0 | 0 |
| .................................................................................................................................................TCATGTGTCGACAACATACTTTTCGTGT.............................................................................. | 28 | 0 | 20 | 0.20 | 4 | 3 | 1 | 0 | 0 |
| .......................................................................................................................................................................TCGGGTTTTAACATTCAGAAAT.............................................................. | 22 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GGAGAGTTCATCGACCTCTGTTGAGG....................... | 26 | 0 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................TTGTCGTAATGCAGCCTTTAAGTAT..................................................................................................................... | 25 | 0 | 19 | 0.11 | 2 | 0 | 2 | 0 | 0 |
| ..............................................................................................................TGTCGTAATGCAGCCTTTAAGTATT.................................................................................................................... | 25 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................................................................TGTCGTAATGCAGCCTTTAAGTAT..................................................................................................................... | 24 | 0 | 19 | 0.11 | 2 | 1 | 1 | 0 | 0 |
| ..............................................................................................................................................CATTCATGTGTCGACAACATACTTTTCGT................................................................................ | 29 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................TCATGTGTCGACAACATACTTTTCGTG............................................................................... | 27 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................................CATTCATGTGTCGACAACATACTTT.................................................................................... | 25 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................CATTCATGTGTCGACAACATACTTTT................................................................................... | 26 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................TGAAGTTCTTGTCGTAATGCAGCCTT............................................................................................................................ | 26 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................................ATTCATGTGTCGACAACATACTT..................................................................................... | 23 | 0 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................CTTGTCGTAATGCAGCCTTTAAGTATT.................................................................................................................... | 27 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................TCTTGCCGTAATGCAGCCTTTAAGTAT..................................................................................................................... | 27 | 1 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................GTCGACAACATACTTTTCGTGTTTT........................................................................... | 25 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................ATGTCGTAATGCAGCCTTTAAGTAT..................................................................................................................... | 25 | 1 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................GTGTCGACAACATACTTCTCGTGTT............................................................................. | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................TCTTGTCGTAATGCAGCCTTTA.......................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................ACTGAAGTTCTTGTCGTAATGCAGCCT............................................................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................CATGTGTCGACAACATACTTTTCGTGT.............................................................................. | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................TGTGTCGACAACATACTTTTCGTGTTT............................................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................GAAGTTCTTGTCGTAATGCAGCCT............................................................................................................................. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................GTTCTTGTCGTAATGCAGCCTTTAAGT....................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................AAGTTCTTGTCGTAATGCAGCCTTG........................................................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................TGTGTCGACAACATACTTTTCGTGT.............................................................................. | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................GTGTCGACAACATACTTTTCGTGTT............................................................................. | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................GTGTCGACAACATACTTTTCGTGTTT............................................................................ | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................GAAGTTCTTGTCGTAATGCAGCCTT............................................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
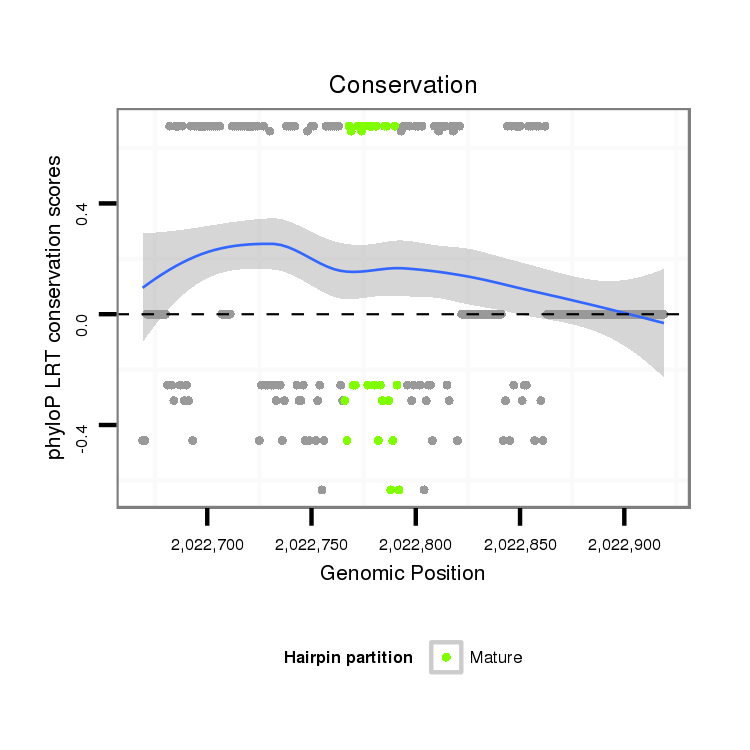
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004961:2022669-2022919 + | dwi_3875 | TTCGACCCCGGGCTAAAAGATAATTTTTTTTTTTATTTTGTAATAATTTAATTTTTTTTCTCACAAAAAAAAAACATTAGCAACTAGTTTAAAAATAATTGACTTCAAGAACAGCATTACGTCGGAAATTCATAAGAGCTATGTAAGTACACAGCTGTTGTATGAAAAGCACAAAATTGTAATTCTTTTTTAT--ATTTGCAGGCCTCTCAAGTAGCTGGAGACAACTCCCGCTTGTGAAGAGCCACAACCTG |
| droPer2 | scaffold_7589:505-665 + | GG----------TTGTAAAAAGTTGTTTTTTTTTATTT-----TAATTTAATTTTTGCTTCCGTTGGCTAAAAATTACCGAAAAAGCGTTAAAAACTTGTGGTTTCAAAAATATTTTTTGTTTCGAAGTATATGACTATGATGTAAAAACAAA--------------------CTAGTATAAACTTTTGTTTGAAA--------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 08:44 AM