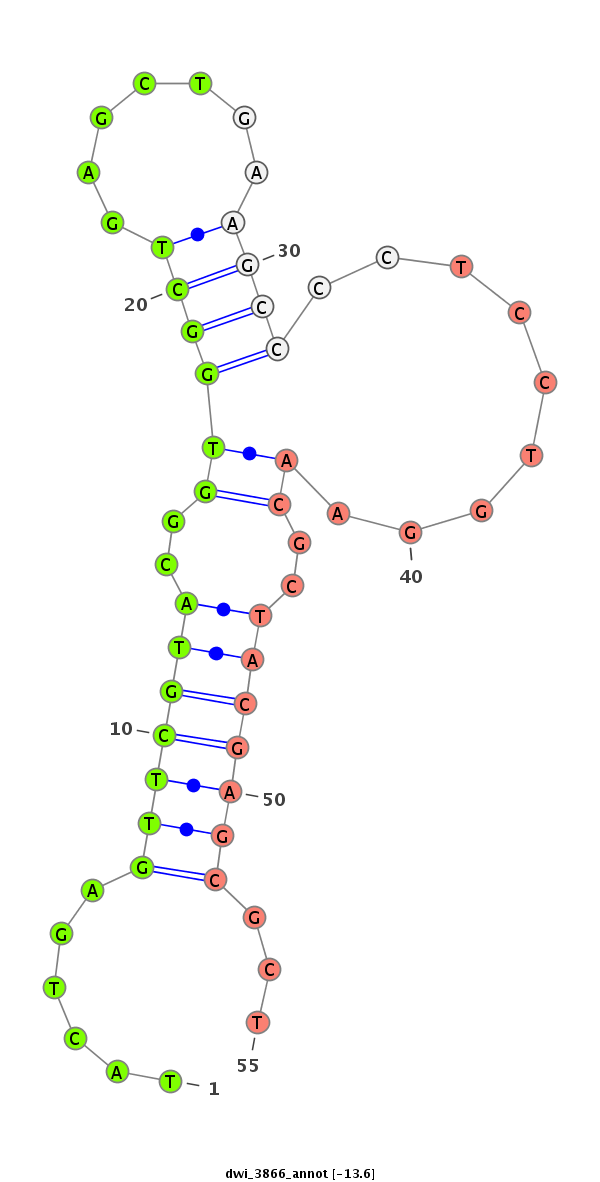
ID:dwi_3866 |
Coordinate:scf2_1100000004961:1021169-1021319 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
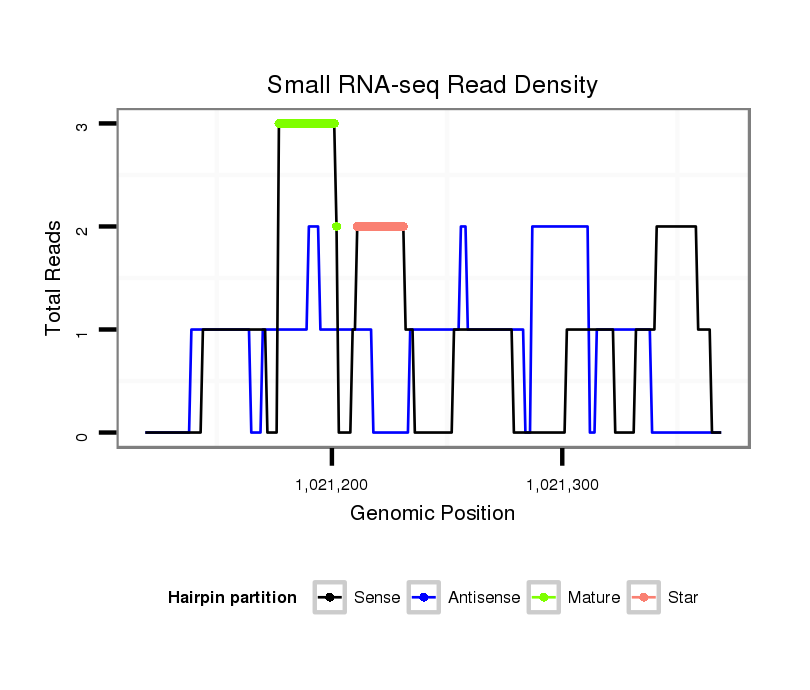
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
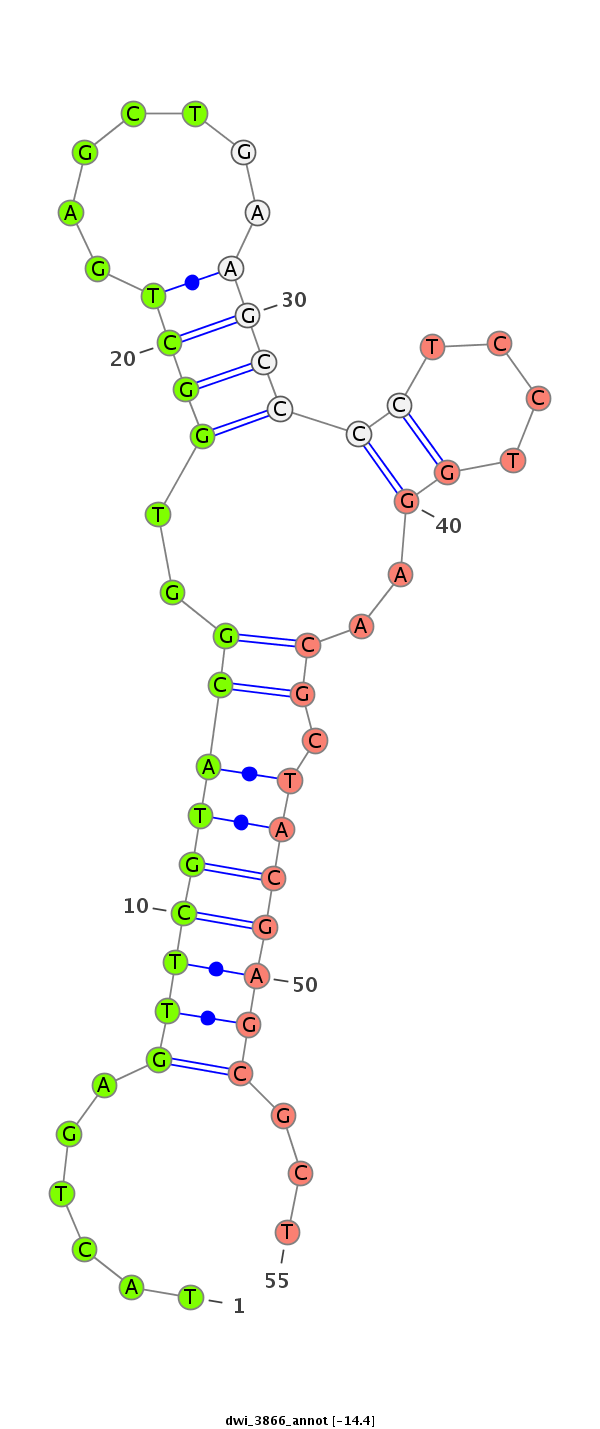
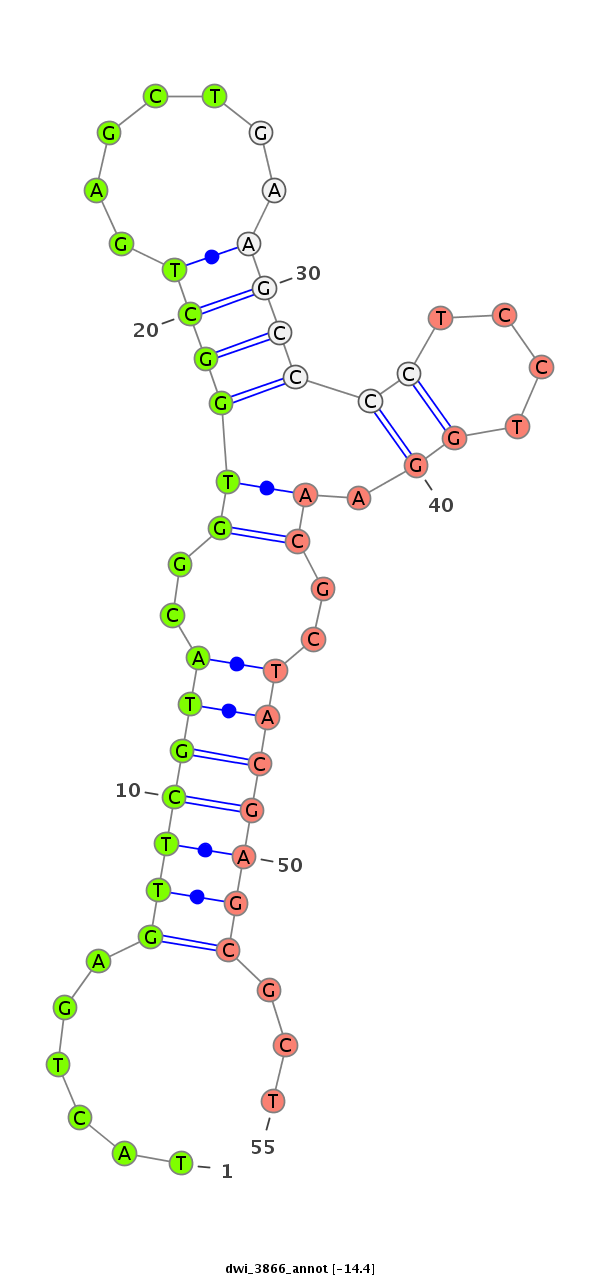
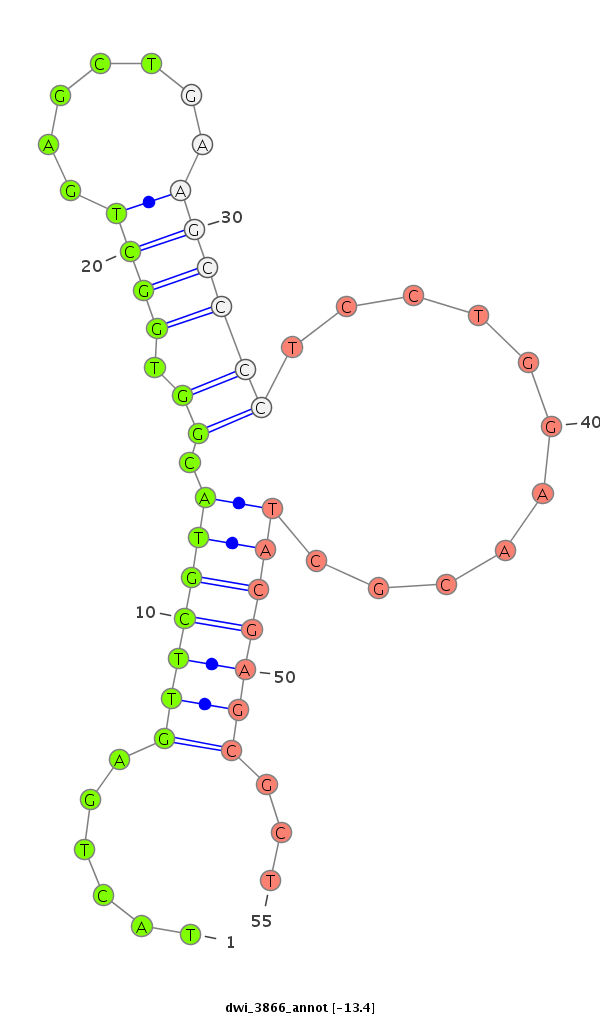
| -14.4 | -14.4 | -13.4 |
 |
 |
 |
CDS [Dwil\GK23429-cds]; exon [dwil_GLEANR_6886:3]; intron [Dwil\GK23429-in]
| Name | Class | Family | Strand |
| Gypsy-12_DWil-I-int | LTR | Gypsy | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCTCGTCGACCTGATCATCGACGGTCTGCGGGACAACTCTACTAGAGTCAGTATGTTGTACTGAGTTCGTACGGTGGCTGAGCTGAAGCCCCTCCTGGAACGCTACGAGCGCTCCCGTGTACAGGCTGCGCCGCCAAGCAAAGGGTCAGTGTAAAGGTGCAGCCAACACCAACTGCCTTGTCGTCCGTGAGCAAATCTTCAGGCCCTGCCACGAGTGAGACCCGATGTTTCAAATGCTCGGGATGGCGGCA **********************************************************......(((((((..((((((.......)))).........))..)))))))...****************************************************************************************************************************************** |
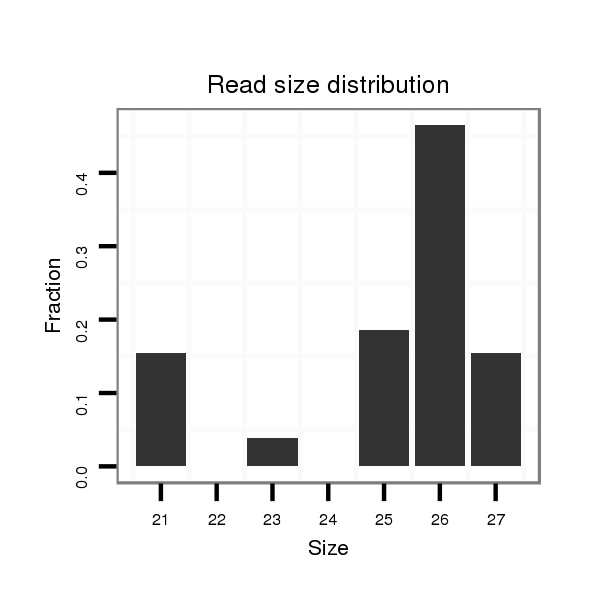
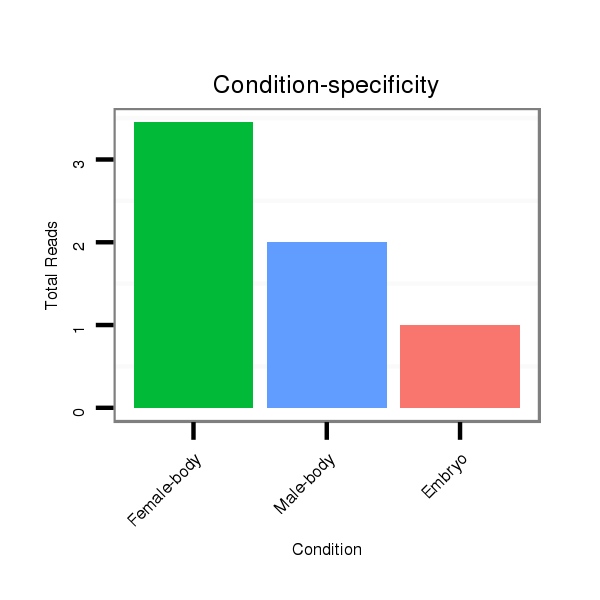
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
V118 embryo |
V119 head |
|---|---|---|---|---|---|---|---|---|---|
| ..........................................................TACTGAGTTCGTACGGTGGCTGAGCT....................................................................................................................................................................... | 26 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 |
| ..........................................................TACTGAGTTCGTACGGTGGCTGAGC........................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TCCGTGAGCAAATCTTCAGGC............................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................TCCTGGAACGCTACGAGCGCT.......................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................................................CCTCCTGGAACGCTACGAGCGCTCCCG...................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................CAAGCAAAGGGTCAGTGTAAAGGTGC........................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| .........................CTGCGGGACAACTCTACTAGAGTCAGTA...................................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................AGTGAGACCCGATGTTTCAAATGCTCG........... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................CGATGTTTCAAATGCTCGGGATGG..... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............ATCATCGACCGTCTGCGT............................................................................................................................................................................................................................ | 18 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................CAAATCTTCAGGCCCTGCCACGA..................................... | 23 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 |
| ..................................................................................................................................................................CCAACACCAACTGCCTTGTCGTC.................................................................. | 23 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ...CGTCGACCTGATCACCTCCG.................................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................AACGCTACGAGCGCTCCCGTGTACA................................................................................................................................ | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................CAAATCTTCAGGCCCTGCCACGAGTGA................................. | 27 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................CAAACATTCCGGCCCTGCCACG...................................... | 22 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................GTGCGGCCTAGCAAAGGGT......................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 |
| .CTCGTCGGCAAGATCATCGA...................................................................................................................................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................................AAAGGAGCGGCCCACACCA................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
CGAGCAGCTGGACTAGTAGCTGCCAGACGCCCTGTTGAGATGATCTCAGTCATACAACATGACTCAAGCATGCCACCGACTCGACTTCGGGGAGGACCTTGCGATGCTCGCGAGGGCACATGTCCGACGCGGCGGTTCGTTTCCCAGTCACATTTCCACGTCGGTTGTGGTTGACGGAACAGCAGGCACTCGTTTAGAAGTCCGGGACGGTGCTCACTCTGGGCTACAAAGTTTACGAGCCCTACCGCCGT
******************************************************************************************************************************************......(((((((..((((((.......)))).........))..)))))))...********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V118 embryo |
V117 male body |
M020 head |
V119 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................GGTTGACGGAACAGCAGGCACTCGT.......................................................... | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................CGTTTCCCAGTCACATTTCCACGTCGGT...................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................GAAGTCCGGGACGGTGCTCACTCT............................... | 24 | 0 | 6 | 1.00 | 6 | 6 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GCACATGTCCGACGCGGCGGTTCGT............................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................ATACAACATGACTCAAGCATGCCAC............................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................TGCCAGACGCCCTGTTGAGATGATCT............................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................ATTGTGGTTGACTGAACAGCAGGCACT............................................................. | 27 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................GCCACCGACTCGACTTCGGGGAGGACCT........................................................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................TGAGATGATCTCAGTTAAA..................................................................................................................................................................................................... | 19 | 2 | 6 | 0.50 | 3 | 0 | 1 | 0 | 0 | 2 |
| ............................................................................................................................................................CACGTCGGTTGTGGTTGACGGAACAG..................................................................... | 26 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................CGTCGGTTGTGGTTGACGGAAC....................................................................... | 22 | 0 | 6 | 0.33 | 2 | 0 | 0 | 0 | 2 | 0 |
| .......................CAGACGCCCTGTTGAGATG................................................................................................................................................................................................................. | 19 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................TGTGGTTGACGGAACAGCAG.................................................................. | 20 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TTGGAAGTCCAGGACGGT........................................ | 18 | 2 | 8 | 0.25 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................TAGAAGTCCGGGACGGTGCTCACTCT............................... | 26 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TTAGAAGTCCGGGACGGTGCTCACT................................. | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................GAAGTCCGGGACGGTGCTCACT................................. | 22 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CCACGTCGGTTGTGGTTGACGGAAC....................................................................... | 25 | 0 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................CGGGACGGTGCTCACTCTGG............................. | 20 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AAGTCCGGGACGGTGCTCACTCT............................... | 23 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................ACAGGAGGTACTCGTTAAGA..................................................... | 20 | 3 | 13 | 0.15 | 2 | 0 | 0 | 0 | 2 | 0 |
| ..................................................................................................................................................................GGTTGTGGTTGACGGAACA...................................................................... | 19 | 0 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....CAGGTGGACTAGTATCTG..................................................................................................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................CATGTATGGCGCGGCGGTT.................................................................................................................. | 19 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TTGGAAGTCCGGGACGGTA....................................... | 19 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TGGGCAAGAAAGTTTACGC............. | 19 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........GACTGGTAGCTGACACACG.............................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TTGGAAGTCCAGGACGGTA....................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004961:1021119-1021369 + | dwi_3866 | GCTCGTCGACCTGATCATCGACGGTCTGCGGGACAACTCTACTAGAGTCAGTATGTTGTACTGAGTTCGTACGGTGGCTGAGCTGAAGCCCCTCCTGGAACGCTACGAGCGCTCCCGTGTACAGGCTGCGCCGCCAAGCAAAGGGTCAGTGTAAAGGTGCAGCCAACACCAACTGCCTTGTCGTCCGTGAGCAAATCTTCAGGCCCTGCCACGAGTGAGACCCGATGTTTCAAATGCTCGGGATGGCGGCA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 05/18/2015 at 08:36 AM