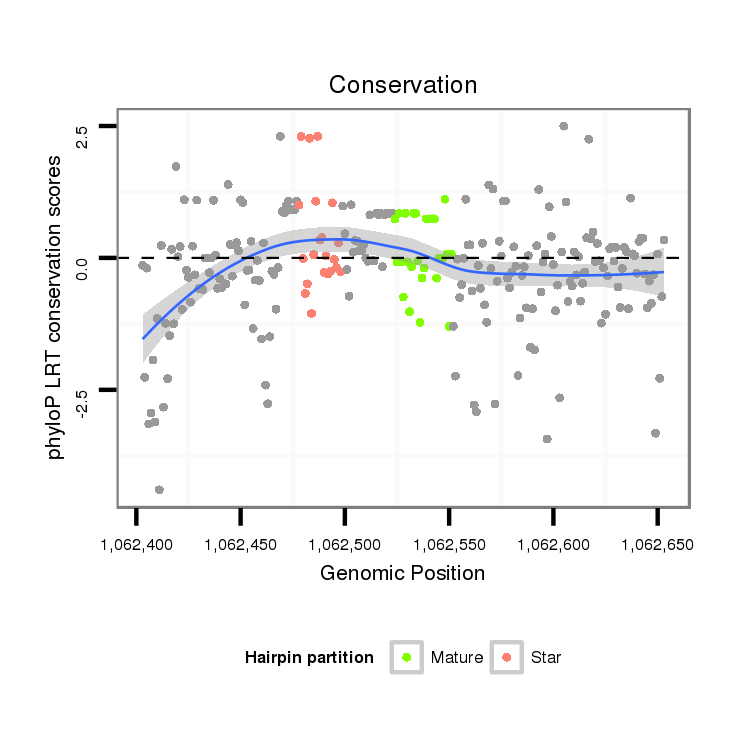
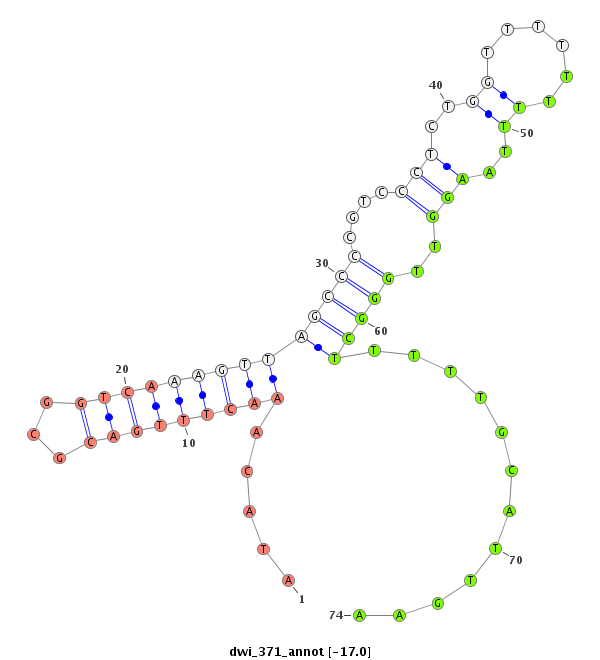
| droWil2 |
scf2_1100000004401:1062403-1062653 + |
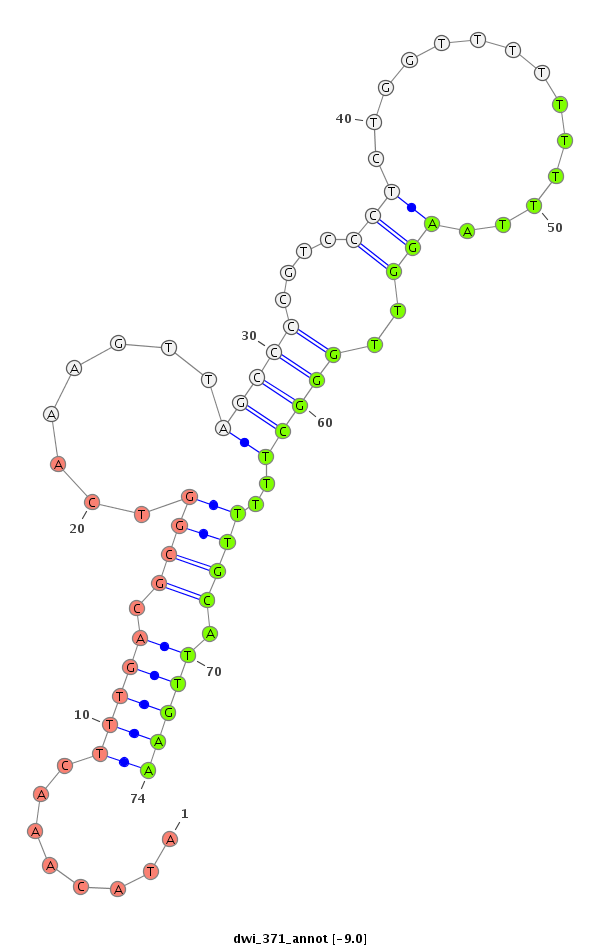
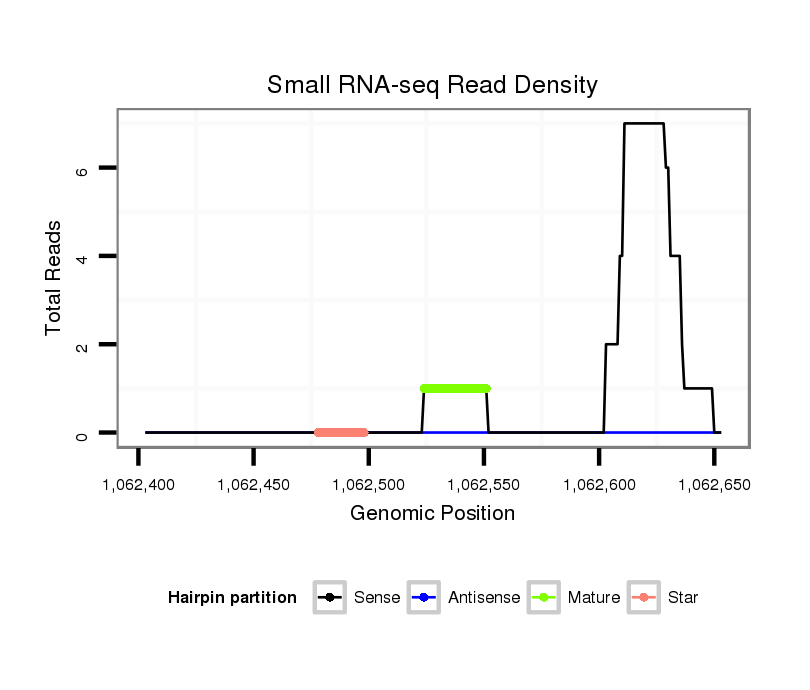
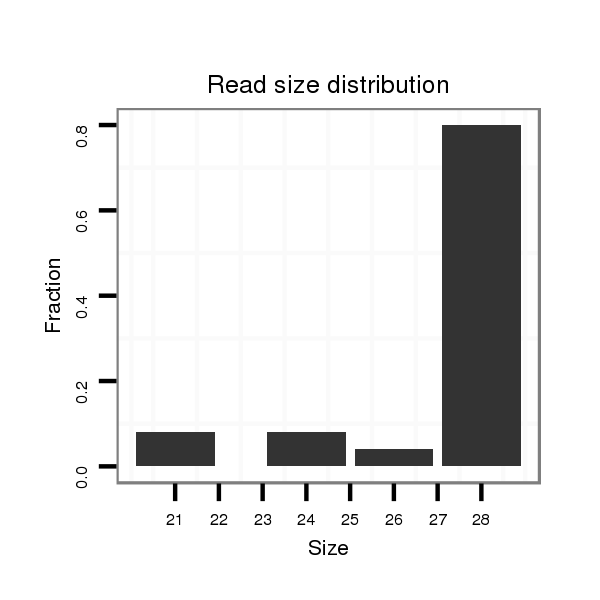
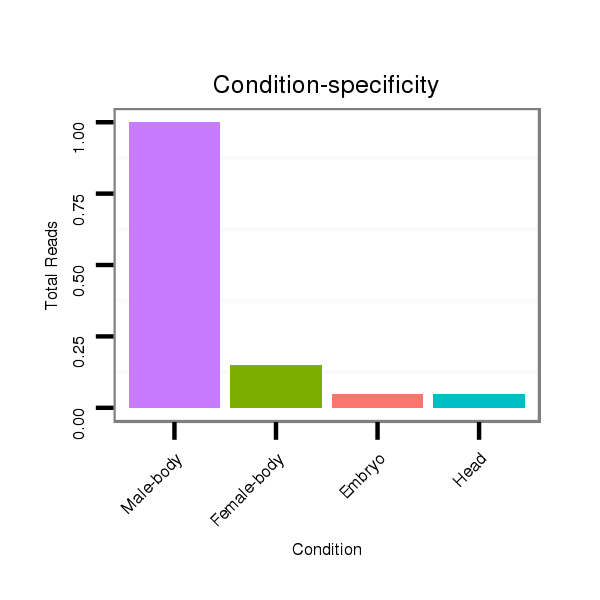
dwi_371 |
TGATCAATA-----------------------TCTTCG--TTATT--TCCT------------------------------AAGCCGTT--CTTTCGCAAACTTA-------------------------------------GAC--------------------CTTTTACA-CA--------------------------------------------------------AAAAACTTGCAAGGGTATACAAACTTTGACGC-GGTCAAAGTTAGCCCCGTCCCTCTGGTTTTTTTTTAAGGTTGG-----GCTTTTTGCAT-----------------------------------------------------------------------------------TGAAATTTTTTTTTCGATTTATCT--------------------------------------TA---------TAAATCCATT------TGA-------------------AT-----C-TCTT---TTTGTTAC-A--GTTTCGATGTA------GAAAAT-----GGCCAA--------------------------------------GGCGCC--CGATCACCGCTTG-----------------------------------------------------------AGGGCGGCT |
| droVir3 |
scaffold_13042:762911-763173 + |
|
TGAACAACT-----------------------TTTTTG--TTA---------------------------------------------------------ATACAGATATCCTAATCATACCCGGCTTC-------------AACTATTCACCCTG-------------TGCT-GAGCATTATTAAAAG-------------------------------G-----------TTGGGTCTGCAAGGGTATTGCATCTTCGGTGT--GCCGAAGATAGCCCT---------------------------------------------TCTTTCTCGTTTTTTTTTTT-TCGTTTTTC------ACGCC-AAATTGA-T-----------------TTTCAGTTA----------------------------------------------------------------------------------------------------------------TTT------TGCAT--GCC--AA---TTGGCTGTTTGGC----TCTTG-----TTGCGC-----CCGTTTCGGATTACCACAGC-------------------GTCGC---CTGCTGCCGCTTG-----------------------------------------------------------CTGCTGGCT |
| droMoj3 |
scaffold_6308:2372346-2372438 + |
|
ATAATCATA-----------------------TTCA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-----------------------------------------ATTTCTTTTC----GTTTTCCCC-------CCATTTAC-A--GTTTCGATGTA------GAAAAT-----GGCCAG--------------------------------------GGCGCC--CGATCACCACTTG-----------------------------------------------------------AGGGCGGCT |
| droGri2 |
scaffold_14830:5655563-5655723 + |
|
TGACCAAAC-----------------------TCTTCA--CTT----------------------------------------------------------------------------------------------------------------A-------------TGTTCAAGAATAATAAATAGGTTCTTAAACTTATAGTAAATTTGAGACAAT-GCCAATTTGTCTCCGGTCTAAGAGGGTGTTTAATCTTCGACAT--GCGTCAGAAAGC----------------------------------------------CTTCTTTCTTTGTTGTTGTTG-----------------C----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTG--------------------------------------GGCTCT--TC----------------------------------------------------------------------------ACA |
| dp5 |
XL_group1e:2752919-2753034 + |
|
TTATATGTA-----------------------CATA-----TATCTCTGTAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAATTCCATATTTTGCTTTGGTTTT-----------------------------------------------------------------------C---------T----T----TATT-------TCATTTAC-A--GTTTCGATGTA------GAAAAT-----GGCCAG--------------------------------------GGCGCT--CGATCACCGCTTG-----------------------------------------------------------AGGGCGGTT |
| droPer2 |
scaffold_2349:4066-4142 + |
|
TTTTTTTAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATTATTATTTTTTCATTTGATTTTACAACTAAAATTAAA---------------------------------------------------A----T---------T----T----TTTT-------TTTTTTAT-T--AATTTGATATA------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droAna3 |
scaffold_13117:4013274-4013494 + |
|
TAATA-ATA-----------------------TAGATGC----------------------------------------------------------------------------------GTGGCGCCACCTATCGGACT-------------GC----------------------------------------------------------------G-----------ACTGAACTGCAAGGGTATATAAACTTCGGCTCCGCCCGAAGTTAGCTTT-CCTTTCTTGTTTTTATATATTTTTAG---------------------------------------------------------------------------------------------------TATTTTTAGTTGAGTATCTGAT------------------TTGGTATTTTATTTAT-----------------------------------------TTTTTTGTTT----GT-----T-TTAT---TTTGTTTT-A--TTTTTTTGTGG-GCAA-GACAAC-----AATTAA--------------------------------------AGCACC--CAA-TACCACTT--------------------------------------------------------------------- |
| droBip1 |
scf7180000396439:55043-55273 - |
|
TATCCGATG-----------------------TTTGCG--ATAT-------------------------------------------------------------------------ATATCC---------------------------------------------------G---------------------------------------------G-----------TTTTAACTGCAAGGGTATGTAAACTTCGGCTCCGCCCGAAGTTAGCTTT-CCTTTCTTGTTTT-ATACATATTTACTTTTATTTTTTTT--------------------------------------------------------------------------------------TATTTATTTTTTTACTTTTAATTTTTACGCTATTATTACT---------------------------------------------------A----T---------TTTTAAT-----T-GCAT---TCTATTAC-A--ATT--ATTGTAAAA---ATAAGT-----GACGGA--------------------------------------CGTGTC--GGATGACAAAATG---------------------------------------------------------CCTGAAGGACT |
| droKik1 |
scf7180000302688:11038-11262 - |
|
TGTTTTGTGTGTGTTTTAAGCATTTAAATTTATA------------------------------------------------------------------ATATAA-------------------------------------------------------------ATAACA-A---------------------------------------------A-----------ACCAATCTGCAAGGGTATACCATTTTCGGCGT--GCCGAAGTTAGC----------------------------------------------TTCCTTTCTTGTTCAATTACA-----------------A------------TATTCTTCATTAAACCCGTATT--------------------------------------------------TTGTTAT---------------CGAATATCTACT-----------------A----T---------TTTTA------TGTAG--ATC--TA---TCG--ATT--ATTGCGTCAA--TGAAGT-----AGCTAA--------------------------------------TGCATT--ATTCTAATGCATA-----------------------------------------------------------AGAGCG--- |
| droFic1 |
scf7180000454040:432367-432586 - |
|
TTAATAGAA-----------------------AAGTCG--T------------------------------CATAGCTTCGAGGTTGTTAGCCTTTATGTATTTT-------------------------------------G------------------------------------------------------------------------------TTCGATTTGTTTGATGACTGCAAGGGTATATAAGCTTCGGCTT--GCCGAAGCTAGC----------------------------------------------TTCCTTTCTTGTTTGTCCTTCTGTTAG----------------------------------------------------------------------------------------------------------------------GGAATATT---------CTAGCTTAATTTTATT-T---------C----AT----TAT-------ATTCTAAT-T--TATTTAGT----TCAAG-----TGTTGC-----TTATTTCAAAATGGCCTA---------------------TACATT--CGAT----------------------------------------------------------------------------- |
| droEle1 |
scf7180000491268:1377654-1377898 - |
|
AAAAATATA-----------------------TAAAA----TATCTATCAAATAATTGAGCTGCAAATCATCATAGCTTCAATG-----------------------------------------------------------------------------------------------------------------------------------------TTTAATTCTTGCAATAGCTGCAAGGGTATATCAACTTCGGCTT--GCCGAAATTTGC----------------------------------------------TTTTTTTCTTGTTAAATCGTG-----------------C------------TTTTCTTCA------------------------------------------------------------------------------TGATTTTTA---------TAAACTCAGCTTTTGTGGCTTCTC-----TGTTT--------------------GTTTTTCC-A--TTTACACTGCG------AAAAAT-------TTGA---------------------------TCTACGATCAAAGCGCCTCCGTTCAGC------------------------------------------------------------------TGTTTGA |
| droRho1 |
scf7180000776685:1542-1770 - |
|
TTATGAGTG-----------------------TTAAGG--ATGCA--TCAT------------------------------ATGAATAA--TTTTAGAGAATATAA-------------------------------------------------------------TTTACA-G---------------------------------------------------------AATTTTTTGCAAGGGTATATCAACTTCGGTTCCGCTTGAAGTTAGCTTT---------------------------------------------CCTTTCTTGTTTTTATTTTG-TTTTTGTTC------CTTGG-GATTTG-----------------------------------------------------------------------------------------------GTAATATT---------TAAGTATTATGGAATT-A---------C-------------------------------------------------------------------GTTTTTAATTAAATTAGAATCGGACATTTAGCATATAGCTGCCATAGAAAAGA------------------------------------------------------------------TGGCAAA |
| droBia1 |
scf7180000301448:1530-1762 + |
|
TTTTTAATA-----------------------CCTCTGAAG---------------TCAGCAACAATCCTTAAAAGTTTCA-------------------------------------------------------------------------------------CATGGTG-TT---------------------------ACTAAAG-----------TTGATTATTTCTTATAACTGCAAGGATATACAAACTTCGGCTG--GCCAAAATTAAA----------------------------------------------ATCCTTTCTTGTTTTTTAATATATT--AGA--CAATTC----GTAAAATAT------------------------------------------------------------------------TTCGGATTTTATAAAT-----------------------------------------ATTTTTTAAA----AC-----T-TTAT---ATTTTTAA-A--GTTTTTATCTG------TCAAAA-----CTTTAA--------------------------------------GGTGC----GATCAC-------------------------------------------------------------------------- |
| droTak1 |
scf7180000415188:4183-4395 + |
|
AAATTAACA-----------------------TTTTTGCGA---------------TTTGTA-------------------A-----------------------A-------------------------------------------------------------TCAATA-G---------------------------------------------G-----------AATTATCTGCAAGGGTATATAAGCTTCGGCTG--GCCGAAGCTAGC----------------------------------------------TTCCTTTCTTGTTTTTTAATGAAAG--GTC--CGATTA----AAAAAATGT-TTTCTACA----------TTTCGGTT-------------------------------------------------------------------AA---------TAAACACAAC------CAAAT--T---------T----C----A-----------TTGACAC-T--TTTTTGAAATC------TTTAAA-----GGTGTG--------------------------------------GGCGCC--AGA-CACCTTTTA-----------------------------------------------------------AAATCG--- |
| droEug1 |
scf7180000408825:29515-29621 - |
|
AAAATTTTC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTAGAAAATTCT--------------------------------------CA---------CTAATTAAATTTTT---GT-------------------AA-----C-TAAT---TTTATTTC-A--GTTTCGATGTA------GAAAAT-----GGCCAG--------------------------------------GGCGCT--CGATCACCGCTCG-----------------------------------------------------------AGGGCGGCT |
| dm3 |
chrX:3215062-3215123 + |
|
TTATCGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTC-A--GTTTCGATGTA------GAAAAT-----GGCCAG--------------------------------------GGCGCT--AGATCACCGCTCG-----------------------------------------------------------AGGGCGGCT |
| droSim2 |
x:2969374-2969491 + |
|
TCGTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TC-A--GTTTCGATGTA------GAAAAT-----GGCCAG--------------------------------------GGCGCT--AGATCACCGCTCATCCGCAGTGAAGGCCAGATGTCTCGCCCCACGGATGAACTTTGTGATCGACTGAAAGCCGCGGGGTAT |
| droSec2 |
scaffold_10:2921428-2921532 + |
|
TTAATTGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTGCAAAAGCATGCTAATCTGATTTT--CTCAACACTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCGTTTC-A--GTTTCGATGTA------GAAAAT-----GGCCAG--------------------------------------GGCGCT--AGATCACCGCTCG-----------------------------------------------------------AGGGCGGCT |
| droYak3 |
3L:21795791-21795905 + |
|
CAAGTGTTG-----------------------TTTTTTT--------------------------------------------------------------------------------ATTC-------------GGTAT-------------GCAAATGTTTTCTTTTGTG-TA--------------------------------------------------------ACATTACTGCAAGGGTATATAAGCTTCGGCAT--GCCAAGTCTGGC----------------------------------------------TTCTTTCCTTTTTTTTT-TAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTTG--- |
| droEre2 |
scaffold_4644:17630-17835 - |
|
TAAAAGAAA-----------------------TCAA--------------------------------------AGTTTCGTTGTTTTTTATTTATAAT-------------TGTTCATATTC-------------GGTATAGAC--------------TGCTTTTTTTAATA-TA--------------------------------------------------------TAATTATTGCATGGGTATAT--GTTTCACCTT--GTCAAAGTTTTA----------------------------------------------CATCGATCCTGTTTTTTTTTTG-TTTAAGACC------TTTCC-AATTTGA-A-----------------TAGCAAATAATTT----------------------------------------CTCG-----------TGATTTTTA---------T---------------------------------------------------------------------------------------------------------------------------------------------------ATTAA-ACTTT-----------------------------------------------------------GAATATCTT |