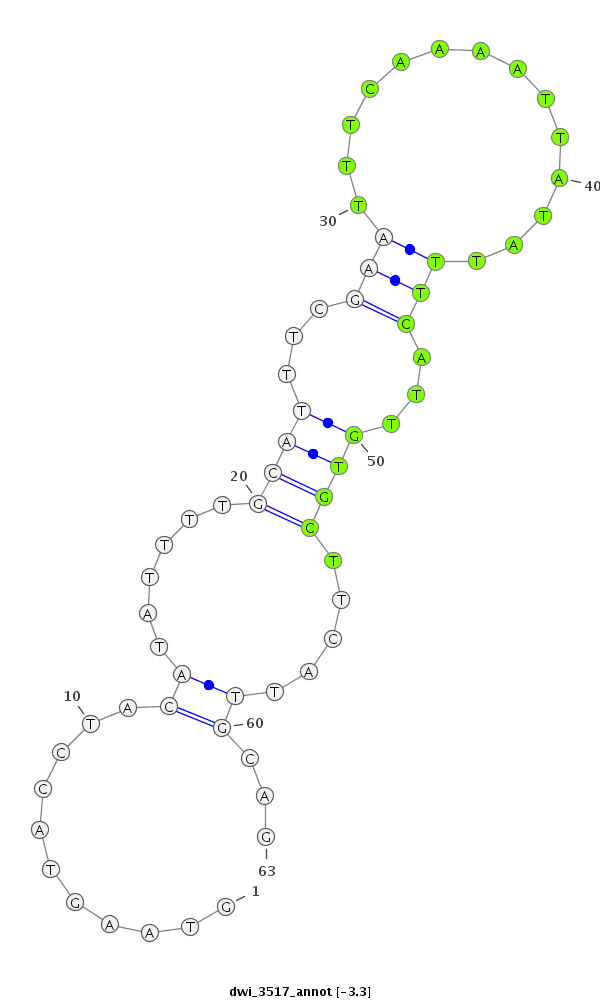
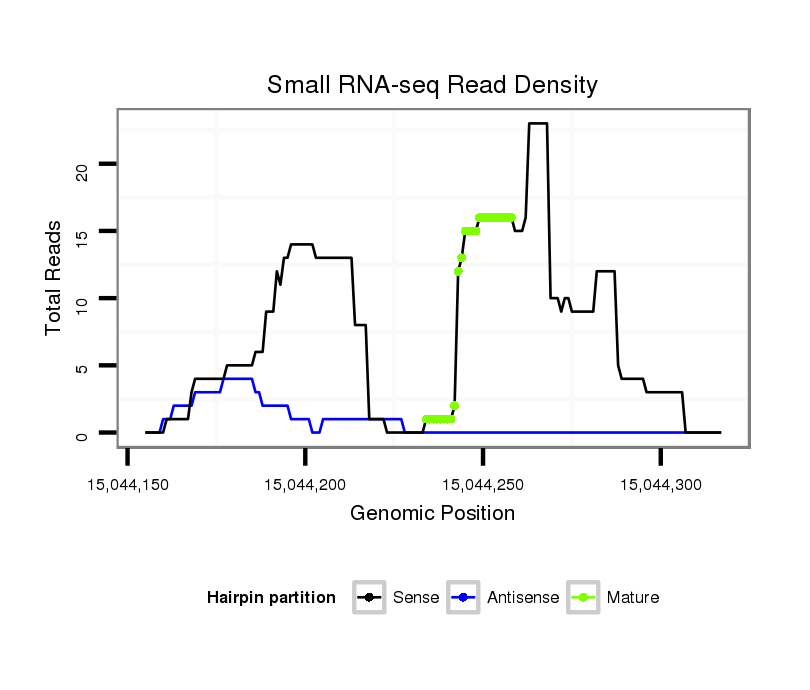

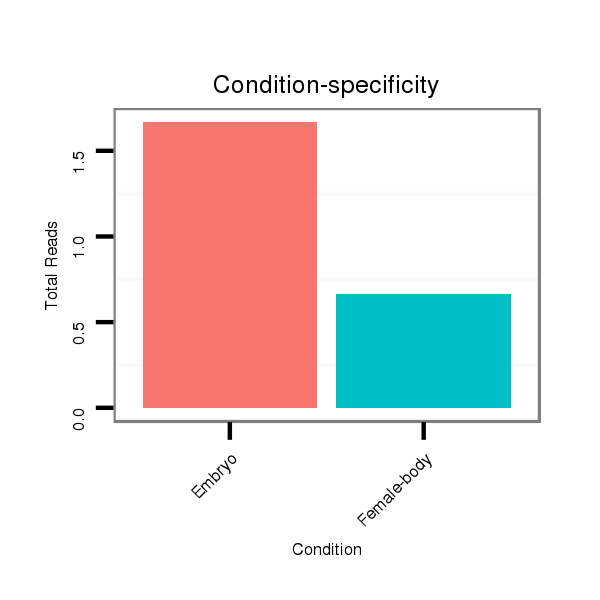
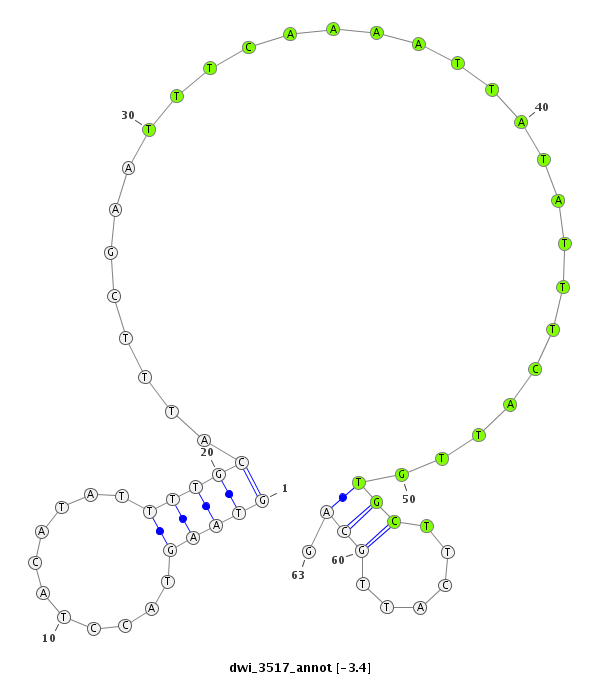
ID:dwi_3517 |
Coordinate:scf2_1100000004943:15044205-15044267 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -3.4 | -3.1 | -3.0 |
 |
 |
 |
exon [dwil_GLEANR_14737:2]; CDS [Dwil\GK14419-cds]; CDS [Dwil\GK14419-cds]; exon [dwil_GLEANR_14737:3]; intron [Dwil\GK14419-in]
No Repeatable elements found
| ##################################################---------------------------------------------------------------################################################## ACGCAAAAAGCGTTTGTCCAAAAAGCGAGCAAGATACTTCATCTGAGCTTGTAAGTACCTACATATTTTGCATTTCGAATTTCAAAATTATATTTCATTGTGCTTCATTGCAGCTGAACAATTTTGATGAAGTTGCTGACATCTTAATGCAAGTGCCCATACC **************************************************...........((......((((...(((..............)))...)))).....))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V118 embryo |
M020 head |
|---|---|---|---|---|---|---|---|---|
| ........................................................................................TATATTTCATTGTGCTTCATTGCAGC................................................. | 26 | 0 | 3 | 10.67 | 32 | 28 | 4 | 0 |
| ............................................................................................................TGCAGCTGAACAATTTTGATGAAGT.............................. | 25 | 0 | 1 | 6.00 | 6 | 3 | 3 | 0 |
| ..................................TACTTCATCTGAGCTTGTAAGTACC........................................................................................................ | 25 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 |
| ...............................................................................................................................TGAAGTTGCTGACATCTTAATGCAA........... | 25 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 |
| .....................................TTCATCTGAGCTTGTAAGTACCTACA.................................................................................................... | 26 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 |
| .............TTGTCCAAAAAGCGAGCAAGATACT............................................................................................................................. | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ......................................TCATCTGAGCTTGTAAGTACCTACA.................................................................................................... | 25 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 |
| .......................................CATCTGAGCTTGTAAGTACCTACA.................................................................................................... | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ..........................................................................................TATTTCATTGTGCTTCATTGCAGC................................................. | 24 | 0 | 3 | 1.67 | 5 | 5 | 0 | 0 |
| .......................................................................................TTATATTTCATTGTGCTTCATTGCAGC................................................. | 27 | 0 | 3 | 1.33 | 4 | 3 | 1 | 0 |
| .........................................................................................................CATTTCAGCTGAACAATTTTGATG.................................. | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................................TCTGAGCTTGTAAGTACCTACATATTT............................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................................................................................ATATTTCATTGTGCTTCATTGCAGC................................................. | 25 | 0 | 3 | 1.00 | 3 | 3 | 0 | 0 |
| ...........................................................................................................TTGCAGCTGAACAATTTTGATGAAGT.............................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ......................................................................................................................CAATTTTGATGAAGTTGCTGACA...................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............................................................................................TCATTGTGCTTCATTGCAGCTGAACA........................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ............................................................................................................TGCAGCTGAACAATTTTGAAA.................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .......................AGCGAGCAAGATACTTCATCTGAGC................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..............TGTCCAAAAAGCGAGCAAGATACT............................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ............................................................................................................TGCAGCTGAACAATTTTGATGAAGTT............................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ......AAAGCGTTTGTCCAAAAAGCGAGCAAGA................................................................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................................................................................TATTTCATTGTGCTTCATTGCAGCTGA.............................................. | 27 | 0 | 3 | 1.00 | 3 | 1 | 2 | 0 |
| ...............................................................................TTTCAAAATTATATTTCATTGTGCT........................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ...............................AGATACTTCATCTGAGCTTGTAAGTACC........................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ............................................................................................................TGCAGCTGAACAATTTTGATGAAGTTA............................ | 27 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................................................................................TTATATTTCATTGTGCTTCATTGC.................................................... | 24 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| .......................................................................................TTATATTTCATTCTGCTTCATTGCA................................................... | 25 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ........................................................................................TATATTTCATTGTGCTTCATTGCAGCT................................................ | 27 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...........................................................................................ATTTCATTGTGCTTCATTGCAGC................................................. | 23 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...............................................................................................CATTGTGCTTCATTGCAGC................................................. | 19 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ..........................................................................................TATTTCATTGTGCTTCATTGCAGCT................................................ | 25 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| .......................................................................................TTATATTTCATGGTGCTTCATTGCAGC................................................. | 27 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| .............................................................................................TTCATTGTGCTTCATTGCAGCTG............................................... | 23 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ..........................................................................................TATTTCATTGTGCTTCATTGCAGCA................................................ | 25 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ..........................................................................................TATTTCATTGTGCTTCATTGCAGCTG............................................... | 26 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| .........................................................................................ATATTTCATTGTGCTTCATTGCAG.................................................. | 24 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ........................................................................................TATATTTCATTGTGCTTCAT....................................................... | 20 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ..........................................................................................TATTTCATTGTGCTTCATTGCAG.................................................. | 23 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ..............................................................................................TCATTGTGCTTCATTGCAGCT................................................ | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ........................................................................................TATATTTCATTATGCTTCATTGCAGC................................................. | 26 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| ............................................................................................................................................ATCTTAATGCAAGTGCCC..... | 18 | 0 | 9 | 0.11 | 1 | 1 | 0 | 0 |
| ................................................................................................ATTGTGTTTCATTGGAGC................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 |
|
TGCGTTTTTCGCAAACAGGTTTTTCGCTCGTTCTATGAAGTAGACTCGAACATTCATGGATGTATAAAACGTAAAGCTTAAAGTTTTAATATAAAGTAACACGAAGTAACGTCGACTTGTTAAAACTACTTCAACGACTGTAGAATTACGTTCACGGGTATGG
**************************************************...........((......((((...(((..............)))...)))).....))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
|---|---|---|---|---|---|---|---|
| ..................................................CATTCATGGATGTATAAAACGTA.......................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .....TTTTCGCAAACAGGTTTTTCGCTCGT.................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ......................TTCGCTCGTTCTATGAAGTAGACTC.................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ........TCGCAAACAGGTTTTTCGCTCGTTC.................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ..............ACAGGTTTTTCGCTCGTTCTATGAAGT.......................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .........CGCAAATAGGTGTTTCGC........................................................................................................................................ | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 |
| ..........GCAAACAGGCTTTTCGGTTG..................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 1 |
| ..............................................................................................AGCAACATGAAGTAACATCG................................................. | 20 | 3 | 19 | 0.05 | 1 | 1 | 0 |
| ....................................................................ACGTAAAGCTTCAAATTT............................................................................. | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004943:15044155-15044317 + | dwi_3517 | ACGCAAAAAGCGTTTGTCCAAAAAGCGAGCAAGATACTTCATCTGAGCTTGTAAGTACCTACATATTTTGCATTTCGAATTTCAAAATTATATTTCATTGTGCTTCATTGCAGCTGAACAATTTTGATGAAGTTGCTGACATCTTAATGCAAGTGCCCATACC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 05/18/2015 at 07:16 AM