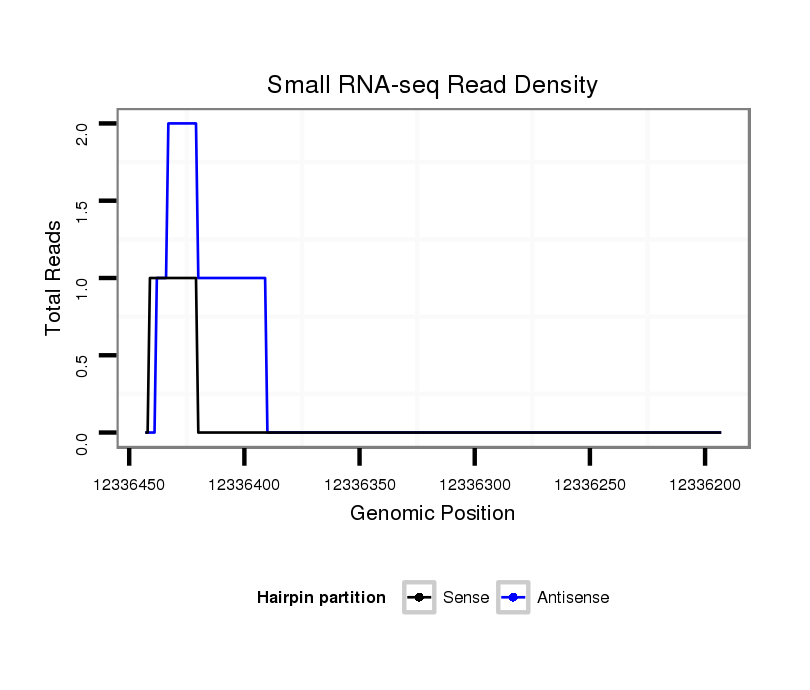
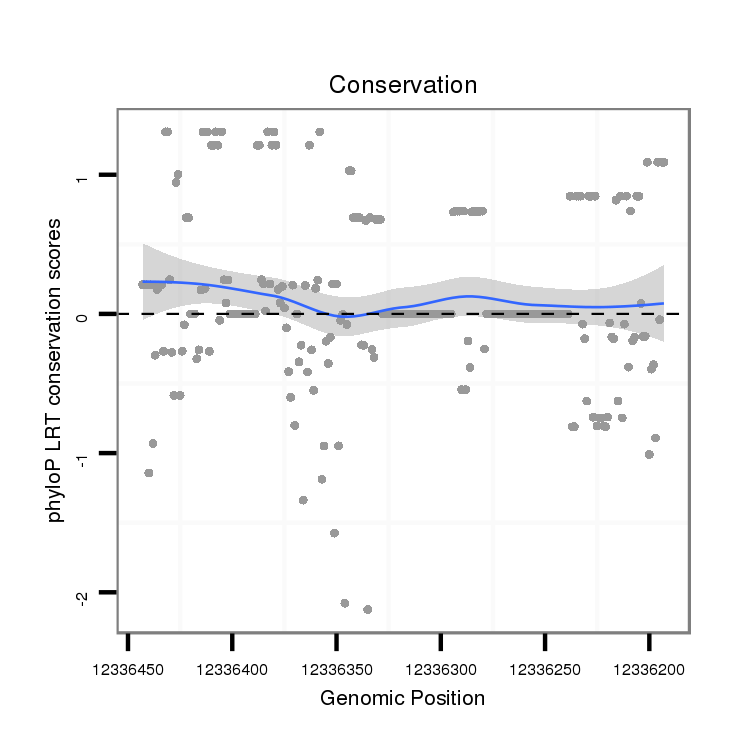
ID:dwi_3450 |
Coordinate:scf2_1100000004943:12336243-12336393 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dwil_GLEANR_13332:1]; CDS [Dwil\GK12991-cds]; intron [Dwil\GK12991-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| -------------------------------###################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATTATTCTAAATTCGACTAAAATTCTGCTTAATGCAGAAGCAGCAGCAAGGTAATGGGAAATGTGACCGCATAAGCACTTGCACTATCTGGTCACACTTAAATATGTCTAGTTATGAGACAATGGCCAGAAAAATTGCCAGAAAGAGCTGAATGTCTTCTTCAATTAATATATAAAATATTTTATTGGATAACAAGTGCTTAAAATTATTTAACAATAAATTAATGTGGATTTCTCCTAAAAACCTCTTTT **************************************************...(((((........)).)))(((((((((.....((((((((..........(((((.......))))).))))))))............((((........)))).((((((.(((((.....)))))..))))))...)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M020 head |
M045 female body |
V118 embryo |
V119 head |
|---|---|---|---|---|---|---|---|---|---|
| .................................................................ACCGCATAACAACTTGGAC....................................................................................................................................................................... | 19 | 3 | 20 | 28.45 | 569 | 296 | 273 | 0 | 0 |
| ..............................................................................................................................................................................AAATATTTTATGGGATAAGAA........................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..TATTCTAAATTCGACTAAAAT.................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................AATATTTTATGGGATAACA......................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................CAAATGCACTATCTGGTGACA........................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................CAATCAATTAATGCGGAT.................... | 18 | 2 | 11 | 0.18 | 2 | 2 | 0 | 0 | 0 |
| ...............................................................................................................................................................................AATATTTTATGGGATAAC.......................................................... | 18 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................AATATTTTATGGGATAACCA........................................................ | 20 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................TTTAATATTTTATGGGATAACAA........................................................ | 23 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................AATATTTTATGGGATAAGAAC....................................................... | 21 | 3 | 17 | 0.12 | 2 | 0 | 0 | 2 | 0 |
| .................................................................................................................................................................................TATTTTATGGGATAACAAC....................................................... | 19 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................AATATTTTATGGGATAAAAA........................................................ | 20 | 2 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................ATATTTTATGGGATAAGAA........................................................ | 19 | 2 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................ACAATCAATTAATGCGGATC................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................AAAATATTTTATGGGATAAA.......................................................... | 20 | 2 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................TATTTTATTGGGGAACAA........................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .................................................................ACCGCATAACGACTTGGAC....................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
TAATAAGATTTAAGCTGATTTTAAGACGAATTACGTCTTCGTCGTCGTTCCATTACCCTTTACACTGGCGTATTCGTGAACGTGATAGACCAGTGTGAATTTATACAGATCAATACTCTGTTACCGGTCTTTTTAACGGTCTTTCTCGACTTACAGAAGAAGTTAATTATATATTTTATAAAATAACCTATTGTTCACGAATTTTAATAAATTGTTATTTAATTACACCTAAAGAGGATTTTTGGAGAAAA
**************************************************...(((((........)).)))(((((((((.....((((((((..........(((((.......))))).))))))))............((((........)))).((((((.(((((.....)))))..))))))...)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
V119 head |
M020 head |
|---|---|---|---|---|---|---|---|---|
| .......ATTTAAGCTGATTTTAAGACGAGT............................................................................................................................................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................CGTCTTCGTCGTCGTTCCAT...................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..........TAAGCTGATTTTAAGACGAATTA.......................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....AGATTTAAGCTGATTTTA.................................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| .....................................................TACCCTTAACACTCGCCTAT.................................................................................................................................................................................. | 20 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004943:12336193-12336443 - | dwi_3450 | ATTATTCTA--------AATTCGACTAAAATTCTG------CTTAATGCAGAAGCAGCAGCAAGGTAATGGGAAATGTGACC-----------------GCATAAGCACTTGCACTATCTGGTCAC--------------------------------------------------------------------------------------------------------------------------------ACTTAAATATGTCTAGTTATGAGACAATGGCCAGAAAAATTGCCAGAAAGAGCTGAATGTCTTCTTCAATTAATATATAAAATATTTTATTGGATAACAAGTGCTTAAAATTATTTAACAATAAATTAATGTGGATTTCTCCTAAA-------------------------------------------------AACCTCTTTT |
| dp5 | 2:14249194-14249311 + | ATTTTGCCA--------ATTTTTTCTTAAAT--------ATTTTTAAGCAGAAAGT-------------GGAGAACGTGAGCACTAGAGCAGTGTGACCGCGGAA-TTATAGTTAAAATATACGGTC-------------------------------------------------------------------------------------------------------------------------CGACCCACAGAAATATACCCA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_0:6190806-6190928 - | ATTTTGCCA--------ATTTTTTCTTAAAT--------ATTTTTAAGCAGAAAGT-------------GGAGAACGTGAGCACTAGAGCAGTGTGACCGCGGAA-TTATAGTTAAAATATACGGTC-------------------------------------------------------------------------------------------------------------------------CGACCCACCGAAATATACCAAAATAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13340:9169278-9169482 + | AAAAAATTA--------ATTTT------------CTAAATTTCTAAAGCAGCAAGT-------------GGAGTACGTGAGC------GCTGTGTGACCAGGCAG-CGAGAGTCAAAATATACCGTTAGAACGCGT---TT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------GAATCTGCGCTTCAAA----------------------------------------TATTTTTCTAATAAGCTTACTGGAACAT---ATAGAAAGAATAAACTCATCTCTAGCATTGTTATATTCATAACATA-AATATAAAAAATATTTT | |
| droBip1 | scf7180000396712:324167-324408 + | ATTCTGGTAAAAAATGAATTTT------------CTAAATTTCTAAAGCAGCAAGT-------------GGAGTACGTGAGC------GCTGTGTGACCAGGCAT-TGGTGGTCAAACAATACTGAAATAAAGCGT---AT------------------------------------------------------------------------------ATATACTGGTATAAAATATACTGTTTTAATGGCGCTTTGAA--------------------------------------------------------------------------------------------------------TTATTTTCCAACATATAAGCTGGGATATATTATAGAAAGCGTAAATTATTCTTTAGAACTATAGTATTCATAAAATTACACAGTAAATACTTTTT | |
| droKik1 | scf7180000302461:1875418-1875643 - | TTTCTACTG--------GCTTTGGCTGCT-----GCGGGAAATTAAGGCAGCAAGT-------------GGAGTACGTGCGT------GCAGTGTGACCAGCTGG-CGCTAGAAAAATCTGGCAGCGATAGCGAGCAGTATGACACATCACGCCAAACAGGCTAATATCAAAATAATTGAAATATCAGCAAACAATCGTGAAATCACAAAATGTCTAGCGGATTCTGAAATAAATTATTTTGTTTTT-------------------------------------------------------------------------------------------------------------------------------------------------------GC-------------------------------------------------CAATTTTATT |
| Species | Read alignment | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||
| droKik1 |
|
Generated: 05/18/2015 at 07:05 AM