| droWil2 |
scf2_1100000004921:357218-357578 - |
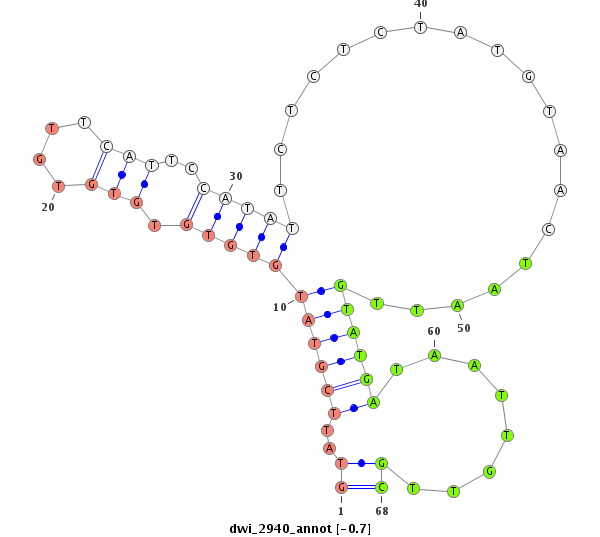
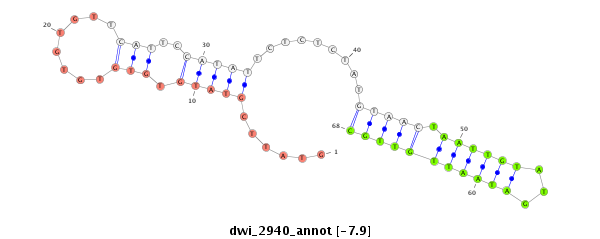
dwi_2940 |
AAAGCGACAGCGGACCCCATCCACCGCCAATCGTGGCCAATAATCCAAAGGTAAG-----CAAAACGGATCAAG------T--TGTGTCA-------AGTTGTCATCGC--------------CCATCTTTATGGC----------------ATTCGTATTCG--TATGTGTGTGTGTGTTCATTC----------------------------------------------------------------------------CATATTCTCTCTAT--------------GTA--------------AC------------------------------------------------------TAATT--------------------------------------------------------------------------------------------------------------------------------GTATGA-------------------TAATTGTTGCTTTATTTTCA-AATGC-------------C-----TA-C------------------------------AACAACAA---C-------------------AACTACAATTAATGCGCCGCTTC--TC---------------TTCTTACCTGCA-----TTCCATA------------AAA--CCAC-CAAAAAAA---CA-------------TTTTTCATCTCAAC--ACATC-TTCTCATC------GCATAACA-ATTTCAACTCTAGAGCAACAAGGTAAAT-GACATAA---TAG---------CTGCTTATC-------------------A-T-CAT-T-TTC---TATAAT---A |
| droVir3 |
scaffold_13047:7917277-7917697 + |
dvi_15951 |
CCGGCGAGCCGGGCCACCACCCACCGCCCATTGTGGCCAACAATCCAAAGGTAAG-----CAAGGTG----------------TTT--------TATCCCTGCCCC--ATGGCCATCGATTGGTGTGCAGCGTGTTTGTCCA-----TATAT------------------------------------------------------------------------------------------------------------------------------------------GTC--------------TA------------------------------------------------------TATT--TATATATAATGTGTTTATATGTAGA----------------------------------GCTATATGTTTT-----------------------GCTTTAATAGTG-------------------------TATAATAACATGTTAAACATTACTCTGTTGCTTTTC-TCTCTCTCTCTCTCTTTGTCTGTCTCGATCTTCA-AT--TGCAACAACAATTGA-TTGCGCGCATCAACAT---T------------AAATACCACCAACGA---T--TGCTGCTTAATG--------------------TATTTGTAACTGCTATCACC---------ATAAA---ACA---TGT------ACA----------------CCCACAA-------CACA-CACACATC-----------------TACATCTGCAGAGCTCCAAGGTAAT---GC-------AGTACTTAGTAATTGCT-GTC-------------------AGTGTACCT-TGTGA----------C |
| droMoj3 |
scaffold_6540:28272950-28273384 - |
dmo_2094 |
CCGGCGAGCCAGGCCACCACCCACCGCCCATTGTGGCCAATAATCCAAAGGTAAG-----CAAGGTG----------------TGTGTC-------TCGTAGCCA----AGGCCTTCGATTGATGTGTAGCGCGTTTGTCCATCTTTGAATAGTTTA--TCTGTGTGTGTGTGTGTGTATGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTAGA---GC------------TAATCGTATTT--AT-------ATGTATT-----------------------GCACAAACTGTGTTAATATCCAAGTATAA-CTTGATTTA-------------------ACTTTGTTGCTTTTTCTCTCTCTCTC------TATCTGTC-----TC-CA-AT--TTCAACAACAATTGATC-GCGC--A-CATCAACATAATAAATAAATATATATACCAACACCGACT-----GCTGCTTTATG--------------------TATATGTAACTGCTATCATC------------TTACAACA---CACAAAC--ACA----------------CTCATACACAC--TCGCC-CACATACT------------------T-TGCTGCAGAGCTCAAAGGTAAT---GC-------AGTAATTAGCACTCGCTCACA-------------------T------------------------A |
| droGri2 |
scaffold_15081:1286297-1286443 + |
|
CATTCGCA---TGT------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTGTGTGTGTGTGTGTAC----------------------------------------------------------------------------TATA-------AAT--------------GTA--------------TG------------------------------------------------------TAATTGTATATTGTACG------TATGTA-----------------------------GATGTATAT-----GC------------ATAT----------GCGA------------------------A-ATTGGCTTA-------------------AAGTGGCAGC-----------------------------------TA-C------------------------------AACAACAA---T-------------------AACAACAATTTCTGC-CTGCGTC--TC---------------TCCT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| dp5 |
2:9719601-9720045 - |
|
CGAGCGAG---GGCTCCCATCCGCCGCCGATAGTGGCCAACAATCCAAAGGTAAG-----CAAGGTG----------------TGT--------GATCGTTGCCTC--CCCACCTTCGAT-GTTGATCCGTGCGTCCATTC----CAGACATATTCG--TTCG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATGTGTACTG------TATGTT-----------------------------AATTAATGTTGTGTGTG--TCATTTGCTGTACTCTGGGG-AG-----------------------AGATTT-CAACACTAA---------------GA--TCTCTCTTGCTTTATCTTCCCTATGT-------------C-----C------TCAAACAACAACAATGCGAC----------ATCGA---C----------------ACTGCCATCGA---CTGTGCTGCTTTTCT--------------------TACCTGCTGCCCCCACCATCATCATCATCATGAACGCACCCCCCACTACA--TGCCACGCCCCACACATCTTTCGTATCTAT--CTATA-CCCACATT------CG--CACATGCCTCAACTGCAGAGCACCAAGGTAAT---GCCTAATGCCTT---------TTGTCTGCCAGTGAGAGTCTCCCCCACAGA-----------------------A |
| droPer2 |
scaffold_0:10760883-10761035 + |
|
CGAGCGAG---GGCTCCCATCCGCCGCCGATAGTGGCCAACAATCCAAAGGTAAG-----CAAGGTG----------------TGT--------GATCGTTGCCTC--CCCACCTTCGAT-GTTGATCCGTGCGTCCATTC----CAGACATATTCG--TTCG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATGTGTACTG------TATGTT-----------------------------AATTAATGTTGTGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droAna3 |
scaffold_13340:19611415-19611808 - |
|
------AA---GGCACCCACCCGCCGCCCATTGTGGCCAACAATCCAAAGGTGGGGCTC---------------------------------------ATTCCAAC--AACTCATTCAACTGTTG--------------------------------------------------------------------------------------------------CCATGAATGTT--------------------------------AACTTT-AGTT---CA---TCAAAGTCA---------T-----------------------------------------------------------------------------------------------ATATATA-----TTATCATATTT--TTAAG-----TGT---CTAA---CTTTGT-TC--AA--------------------------AAACAT-TTTGACTAA-------------------GACATGTTGCTTTATCTTCCTTATGC-------------C-----AA-CC-----AACAACAACAATGTTACTGCGC--TTCATCAT---C-------------------GACATTGACTAATGCGGTGCTTACTCCACATTCGAACCGCCCCACACACCCACA-------------CCA--ACCATAAAAACTCAC-CCAAAAAA--CAC----------------CACA-----------TCA-CACACATCAAAACTCG--CACATGCCTCGACTGCAGAACAACAAGGTAAT---GC-------CAT---------CTGTCCACC-------------------TGGGAT--T-TGTGAGG---AT---T |
| droBip1 |
scf7180000396413:2774381-2774720 + |
|
------AA---GGCTCGCATCCTCCGCCCATTGTGGCCAATAATCCAAAGGTGCG-----GA--TCATAAAAAT------TTG---TTGGAAA-AA------TCATACT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA---------TTTATGT---ATTTCAT--------------------------------------------------AA------------------------------------------------TT--TGACT-----TGT---CTAA---CTTTATTCG--AA---------------------------ATCAT-TTTGACTAA-------------------GATCTGTTGCTTTATCTTCCTTATGC-------------C-----CA-CC-----AACAACAACAATGTTACTGCGC--TTCATCAT---C-------------------AACATTGACTAATGCGGTGCTTTCTCGACATCCAAACCGTCCCAAAAA--CACT-------------CGA--ACCAT-------------AAAAAC--TCA----------------CCCACAT-------CTCA-CACACATTTAAACTCG--CACATGCCTCGACTGCAGAA---CAAGGTAAT---GC-------CAT---------CTTTCCATC-------------------TGAG----------A----------- |
| droKik1 |
scf7180000302705:134561-134719 + |
|
CAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACAA---C-------------------AACATCGACTAATGCGCTGCC--TTAC------------------ACATTTCCA-----TTTCATA------------AA-ACATAT-TCACTCAC--TTA----------------CTCACTGCCAT--ATA-----CACATC------CACATGCACACCTCGACTGCAGAACACTAAGGTAATAAAGC-------CAG---------CTGCTCCGA-------------------G-A-AAA-T-CCA---GGGAAT---A |
| droFic1 |
scf7180000453826:635072-635509 - |
|
CCAGCGAT---GGATCCCACCCACCGCCCATCGTGGCTAACAATCCAAAGGTAAA-----AGCACGTGAACCTCAAAAAGT---GCACCA------------------A--------------AAATGCAT--------CT--------TGT----------------ACCCATTTTTAC------ATCTGGACCATTTCTTATTCGGATTTCGTTTTCACAAAAT--ATCATAAACAATTTAAATAAAATGACCTCGATTCCAAATACTTT-AGT--------------------------------------------------------------------------------------------------------------------------------------TT--------TA--TTAAGTTATG--TT--ACT---------ATCC--AT--------------------------AGACAT-TTGAACTAC-------------------AGTTTCTTGCTTTATCTTCCTTATGC-------------T-----CAACATCTCGAACCACAACAATGCAACTGCGC--AACACCAA---C-------------------AATATCGACTAATGCGCCGCT--TAAC--------------------------A-----CTTCATT------------CCAATCCAT-TGTAAACC--CCA----------------CTCATTTTCAT--CCTCC-CACAAACG------CAAACGCACACATCGACAGCAGAGC---AAGGTAAA---GC-------CAT---------CTGTCCGCC-------------------ATTAAAATT-TCCGA----------- |
| droEle1 |
scf7180000491261:1550179-1550362 + |
|
CCAGCGAC---GGCTCCCACCCACCGCCCATCGTGGCCAACAATCCAAAGGTAAAATAC-AGCCCATGATCCTT------TTG---GTGGATAAAT------TCATCCC--------------GCATTCTTGTGCC----------------ATTTGTA--------TGTA---------------ATAGGTATCAT------------------------------------TAT--------------------------CAACTTTTTATCGT--------------TTC--------------AC------------------------------------------------------TAATT---------------------------------------------------------------------ATT-----------------------TTACAAATTCA--------------------------------------------------------CTTTTTATACA-AGTG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT---T |
| droRho1 |
scf7180000780007:66322-66451 - |
|
CCAGCGAT---GGATCCCACCCACCGCCCATCGTGGCCAATAATCCAAAGGTAAG-----AGCCTATAAACCTC------TTC---GTTGATATGT------TCATCCC--------------GAATCCGTATGCC----------------ATATGTA--------TGTG---------------ATTGGGATCAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-------------G----------------TTTATTC-------------------------------------------------------------------------------------------------------------------------------------------- |
| droBia1 |
scf7180000302075:1870519-1870648 - |
|
C--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACCAA---C-------------------AACATCGACTAATGCGCCGCT---CAC--------------------------A-----CATCATC------------GAAATCCAT-TAAAACAAATTCA----------------CTCATCTCCAT--CCGCC-CTCACATC------CAAACGCA--------CTGCAGAGCACCAAGGTAAA---GC-------CAT---------CTGTCCGCA-------------------GAA----------------------- |
| droTak1 |
scf7180000415700:453960-454351 + |
|
CCAGCGAT---GGCTCCCACCCACCGCCCATCGTGGCCAACAATCCAAAGGTATC-----AGCTTGGGAACCTT--------C---A--TATATGATCATTCCATT--TC------------------CATT------------------------------------------------------------------------------------------AACT--ACCATT--------------------------------TGTA----------CA----------AA---------TTTTTGGAGT-------AAATAACTTGAAATAATATTGTAGATCCTATTTGTAACGCCCC------C---------A----TATA---------------TTTATATGT--------TA--TTATTTTGTATGTA--ACCA---CTTCATTCC--AA--------------------------AGACAT-TCCGACTAA-------------------GATATCTTGCTTTATCTTCCTTATGC-------------T-----CAACAAAACGAACCACAACAATGCTAC--------------------------------------------------TGCGCT-----------------------------------A-----CATCATC------------GAAATCCAT-------------A----------------TTCATTCCCATATCCGCTACCCACATT------CAAACGCA--------CTGCAGAGCACCAAGGTAAAG-AGC-------CAT---------CTGTCTGCA-------------------GAGGAATACC-TA--GA---ATTATA |
| droEug1 |
scf7180000409770:549787-550213 + |
|
CCAGCGAT---GGATCCCACCCACCGCCCATCGTGGCCAATAATCCAAAGGTAACGCCCGCTAAGTC----------------TTT--------TATTCTTTCTTT--TTCCCATTTC----------TTTT------------------------------------------------------------------------------------------TTCAAGACCATT--------------------------------AGTTTC-AGTT---CACAATTAAAGTAATAACCCTCATT--TA-----------TTTTAATTTGA---------------------------------------------AATATATTTATA---------------CATTTATGT--------TA--TTAAGTTGTATATA--ACCA---CTTCATCCC--AA--------------------------TGACAT-TCCGACTAA-------------------GATCTCTTGCTTTATCTTCCTTATGC-------------T-----CAACAACTCGATCCACAACAATGCTAC-GCGC--CTCACCAA---C-------------------AACATCGAATAATGCGCTGCTG---AC--------------------------C-----CATCAAC------------AATATTCAT----AAAAA--CCG----------------CTCATTCCCAT--CCGCC-CTCATACCC-----CAAACGCACATATTGACCGCAGAGT---AAGGTAAA---GC-------CAT---------CTGTCTGCA-------------------GAGGAAATTC-CA--TA---ATCA-G |
| dm3 |
chr3R:23080404-23080768 + |
|
CCGGCGAT---GGCTCCCATCCGCCACCCATCGTGGCCAACAATCCAAAGGTAAG-----CGAAACGGAGCCCT------TGG---GTGGATATTC------TCATCGC--------------GAATCCTTGTGTCTGTCTA------A--------------------------------------------------------------------------------------------------------------------------------CTT---------------------------------A--------------------------------------------------------ACGTA-----------------GAAGTCCATACAAA---------------------------TTGTG--ATAT---CTTCATCCC--AAAAA------------------------AAATT-TCCAACTAA-------------------GATATCTTGCTTTATCTTTCTTATGC-------------T-----CCACCACTTGAACCACTACAATGCAAC-GCGC--CTCACCAA---C-------------------AACATCGACTAATGCGCTGCTCTTTGC--------------------------A-----CATCATC------------AAAATCCAT-TCAAAAAC--CCA----------------TTCATTTCCAT--GCGCC-CTTACATC------CAACCGCACGTTTCGAATGCAGAGCCACAAGGTAAA---GC-------CAT---------CTGTCTGCG-------------------GGA----------------------- |
| droSim2 |
3r:22522108-22522469 + |
|
CCGGCGAT---GGCTCCCATCCGCCGCCCATCGTGGCCAACAATCCAAAGGTAAG-----CGAAACGGAGCCCTAGGGTTA---GGGTCG------------------A--------------GAATCCTTGTGTCTGCCTA------A--------------------------------------------------------------------------------------------------------------------------------CAT---------------------------------AGTCCTT--------------------------------------------------AAGTG-----------------G--GTCCTTACAAT---------------------------GTGTG--ATAT---CTTCATCCC--AA--------------------------TGACATTTCCAACTAA-------------------GATATCTTGCTTTATCTTCCGTATGC-------------T-----CCACCACTTGAACCACTACAATGCAAC-GCGC--CTCACCAA---C-------------------AACATCGACTAATGCGCTGCTATTTGC--------------------------A-----CATCATC------------AAAATCCAT-TCAAAAAA--CCA----------------TTCATATTCAT--GCGCC-CTCACATC------CAAACGCAAGTTTCGAATGCAGAGCCACAAGGTAAA---GC-------CAT---------CTGTCTGCG-------------------GGA----------------------- |
| droSec2 |
scaffold_13:1834645-1835021 + |
|
CCGGCGAT---GGCTCCCATCCGCCGCCCATCGTGGCCAACAATCCAAAGGTAAG-----CGAAACGGAGCCCTAGGGTTA---GGGTCG------------------A--------------GAATCCTTGTGTCTGTCTA------A--------------------------------------------------------------------------------------------------------------------------------CAT---------------------------------AGTCCTT--------------------------------------------------AAGTG-----------------G--GTCCTTACAAT---------------------------GTGTG--ATAT---CTTCATCCC--AA--------------------------TGACATTTCCAACTAA-------------------GATATCTTGCTTTATCTTCCGTATGC-------------T-----CAACCACTTAAACCACTACAATGCAAC-GCGC--CTCACCAA---C-------------------AACATCGACTAATGCGCTGCTATTTGC--------------------------A-----CATCATC------------AAAATCCAT-TCAAAAAA--CCA----------------TTCATATTCAT--GCGCC-CTCACATC------CAAACGCAAGTTTCGAATGCAGAGCCACAAGGTAAA---GC-------CAT---------CTGTCTGCG-------------------GGAGAATTTC-CA--GA---ATC--A |
| droYak3 |
3R:23413057-23413433 - |
|
CCAGCGAC---GGCTCCCACCCGCCGCCCATCGTGGCCAACAATCCAAAGGTAAA-----CGAACTGGAACCCA------CAG---TCGGATATGT------TCATCTT--------------AAATTCTTGTGTCTGTCTA------G------------------------------------------------------------------------------------------------------------------------------CACAT---------------------------------AG-----------------------------------------------------------A-------------------AGTCCTTACCATCTATAT--------TT--TTAAGCTATGTGTT--ACCA---CTTCACCCC--AA--------------------------AAACAT-TCCAACTAA-------------------GATATCTTGCTTTATCTTCCTTATGC-------------T-----CCACCACTTGAACCACAACAATGCAAC-GCGC--CTCACCAA---C-------------------AACATCGACTAATGCGCTGCC--TTAC--------------------------A-----CATCATC------------GAACTCCAT-TCAAAAAC--CCA----------------TTCATTTCCAT--GCGCC-CTCACATC------CAAACGCACATTTCGACTGCAGAGCAACAAGGTAAA---GC-------CAT---------CTGTCTGCA-------------------GGAG----------G----------- |
| droEre2 |
scaffold_4820:4953544-4953906 - |
|
CCGGCGAT---GGCTCCCACCCGCCGCCCATCGTGGCCAACAATCCAAAGGTAAA-----CGAACTGGAAGCCT------CGG---GTGGACATGT------TCATCTC--------------AAATCCCTGTGTCTATCTA------G--------------------------------------------------------------------------------------------------------------------------------CAT---------------------------------A-----------------------------------------------------------TA-------------------AGTCCCTACAATGTATAG--------TT--TTAAGCTATGTGTT--ACCA---CTTCACCCC--AA--------------------------AGACAT-GCGATCTAA-------------------GAAATCTTGCTTTATCTTTCTTATG------------------------------ATCCACTACAATGCAAC-GCGC--CTCACCAC---C-------------------AACATCGACTAATGCGCTGCC--TTAC--------------------------A-----CATCATC------------AAACTCCAT-TCAAAAAC--CCA----------------TTCATTTCCAT--GCGCC-CTCACATT------CAAACGCACATTTCGACTGCAGAGCAACAAGGTAAA---GC-------CAT---------CTGTCTGCA-------------------GGAG----------G----------- |