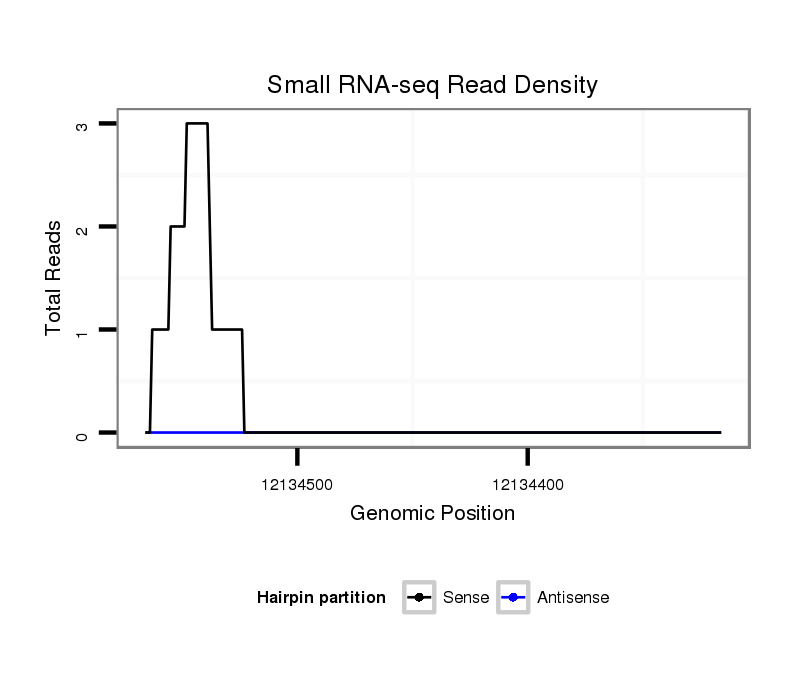
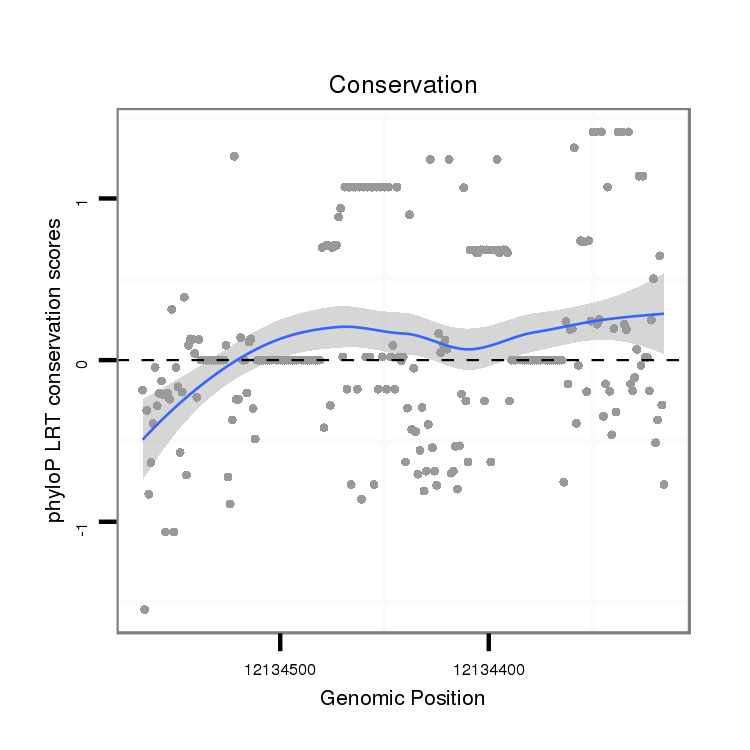
ID:dwi_2912 |
Coordinate:scf2_1100000004909:12134366-12134516 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dwil\GK24914-cds]; exon [dwil_GLEANR_9086:5]; intron [Dwil\GK24914-in]; Antisense to intron [Dwil\GK10242-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TTGCACCGATGGTTCCGACGAAGATCCTCAAATGTGTGCAGCCCACCGAGGTTTGTTCTGCTTGTTGGGGAAAGAAAAATCGGGTTCAATTCAACATACACAACATATATACACATACATTCATATTAAGTCTATTCGTGGAAGGGAATGCAAAACGAAACGAATTTGAAAGAAACAACAAAAATTTTCGGATCAAAGTTACGATTGCAATCCCTGAAAGAGGAGGGGAAAAATGTCAAAAGCTTTTGTGC **************************************************((.((((.((..(((((((....(((........)))..)))))))..)).)))).)).........(((((........((((....))))...)))))......((..((((......................))))..)).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
M045 female body |
V117 male body |
|---|---|---|---|---|---|---|---|---|
| ............................CAAATGTGTGCAGCCCACCGAGAGA...................................................................................................................................................................................................... | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................CGAAGATCCTCAAATGTGTGCAGCC................................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...CACCGATGGTTCCGACGAAGATCCT............................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........GTTCCGACGAAGATCCTC.............................................................................................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........................................................................CAGATCGGGTTCAATTCC.............................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................................GAAAGATGTCAAAAGCTGGT.... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
|
AACGTGGCTACCAAGGCTGCTTCTAGGAGTTTACACACGTCGGGTGGCTCCAAACAAGACGAACAACCCCTTTCTTTTTAGCCCAAGTTAAGTTGTATGTGTTGTATATATGTGTATGTAAGTATAATTCAGATAAGCACCTTCCCTTACGTTTTGCTTTGCTTAAACTTTCTTTGTTGTTTTTAAAAGCCTAGTTTCAATGCTAACGTTAGGGACTTTCTCCTCCCCTTTTTACAGTTTTCGAAAACACG
**************************************************((.((((.((..(((((((....(((........)))..)))))))..)).)))).)).........(((((........((((....))))...)))))......((..((((......................))))..)).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................TTTAGAGGCCTAGTTTCGA................................................... | 19 | 3 | 16 | 0.94 | 15 | 14 | 1 |
| ..................................................................................................................................................................................GTCTTTAGAGGCCTAGTTTC..................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004909:12134316-12134566 - | dwi_2912 | TTGCACCGATGGTTCCGACGAAGATCCTCAAATGTGTGCAGCCCACCGAGGTTTGTTCTGCTTGTTGGGGAAAGAAAAATCGGGTTCAATTCAACATACACA-ACATATATACACATACATTCATATTAAGTCTATTCG-TGGAAGGGAATGCAAAACGAAACGAATTTGAAAG--AAACAACAAAAATTTTCGGATCAAAGTTACGATTGCAA---------TCCCTGAAAGA-GGAGGGGAAAAATGTC---------------AAAAGC--TT-TT---GTGC |
| droVir3 | scaffold_13049:23177211-23177283 - | TCAGCA------------------------------------------------------------------------------------------TATATA-ATATATATATATATATATATATCTGAATACAGTTAA-TGTCAGGGCAATTCTAA---------------A-------------------------------------------------------------------------------------------------------GAA---ATCG | |
| droMoj3 | scaffold_6500:10387981-10388041 - | TGTTGCA----CAAAGAGTATT----------------------------------------------------------------CCATACAACATATAAA-ATAAATATAAATATATATATATATA------------------------------------------------------------------------------------------------------------------------------------------------------------AA | |
| droGri2 | scaffold_15110:1748823-1748979 + | AATCAAACGTA-----------------------------------------------------------------------------------CACACACACACACACACACACACACACACACACAGAGAACACGCACTAGAACTAAATGCA---TCAAACGAACTTCAAAGGCAAACG-------------------------GACGTC--GTAGG-TA------GAAAAAGAA-GAGAAAAACTAAAAACAAAAACAAAAACA-----AAAACTT---GTAT | |
| droPer2 | scaffold_8:3367320-3367327 + | TTTT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGGT | |
| droAna3 | scaffold_12916:5269599-5269701 - | GGATATAAAAGTAAAAAAAAAAGTTCTT------------GGGTATTA--TTCAT-------------------------------------------------------------------------------------------------------------------------------------------------------ATTTTCTTTTAAAAGTTTCCTTAAAAAA-TAATCTTAAAAATAT-----------------ATAATATTTCTCTCAGTGT | |
| droEle1 | scf7180000490478:9888-9959 - | TGATGATGATGATGATGATGATGATTAT------------GATTACCG--ACTTT------------------------------------------------------------------------------TTCCAC-TTTTTGGGAAGTAGCCA---------------A-------------------------------------------------------------------------------------------------------GTC---GTGT | |
| droSim2 | 2r:6193078-6193174 + | CAAAAAAGGACCAAAGGGAAAGCAATAA------------C-AAATTG--ATTAA-------------------------------------------------------------------------------------------------------------------------------------------------------AACTTCGAGTAGA-CG------GAAGAAAGG-AAGAAAAGATGAA---------------GAAAGCAAATCTTTCATTTA | |
| droEre2 | scaffold_4929:8952938-8953035 - | CAAAAAAGGACCAAAGGGAAAGCAATAA------------C-AAATTG--ATTAA-------------------------------------------------------------------------------------------------------------------------------------------------------TACTTCGAGTAGA-CG------GAAGAAAGGAAAGAAAAGATGAA---------------GAAAGCAAATCTTTCATTTA |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 05:33 AM