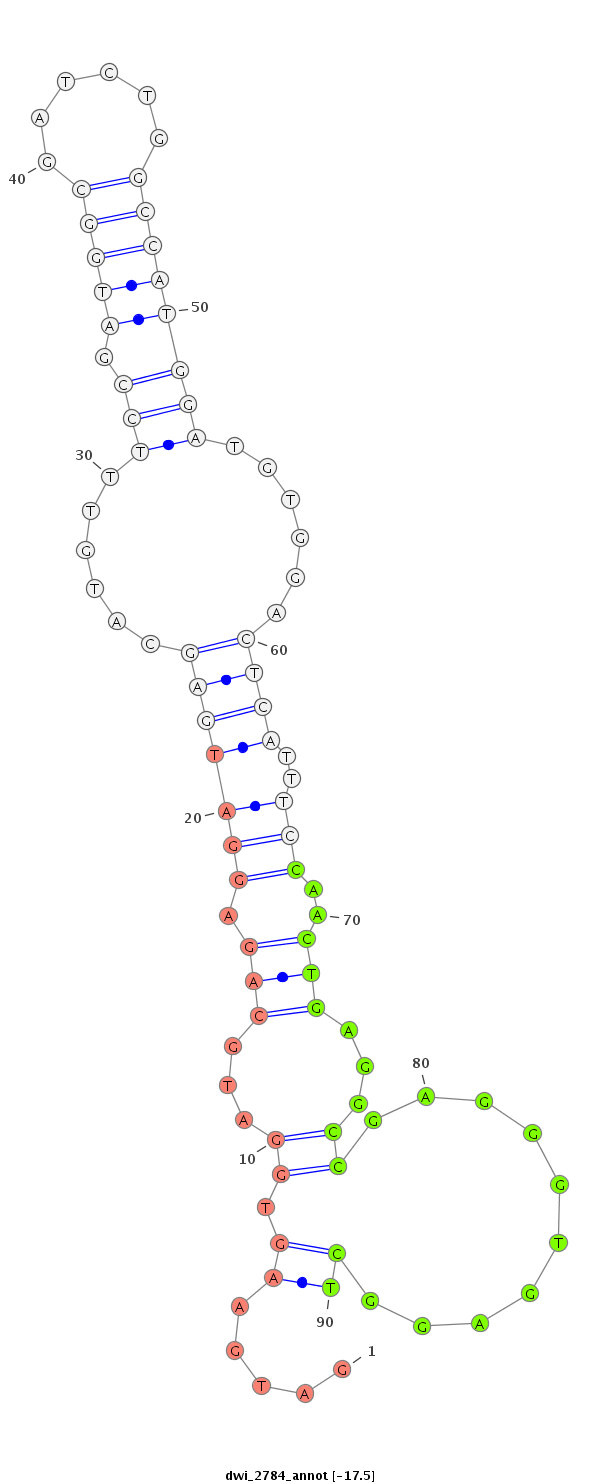
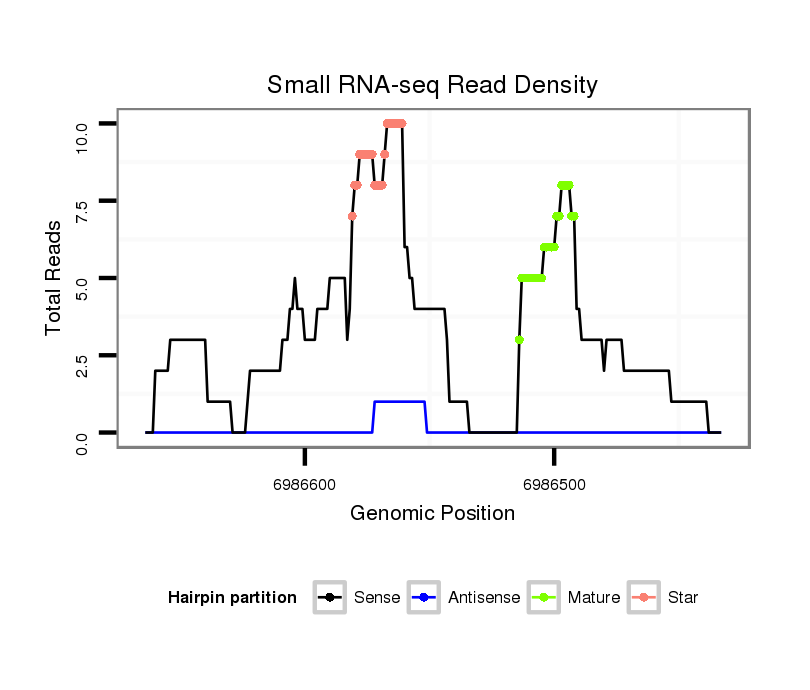
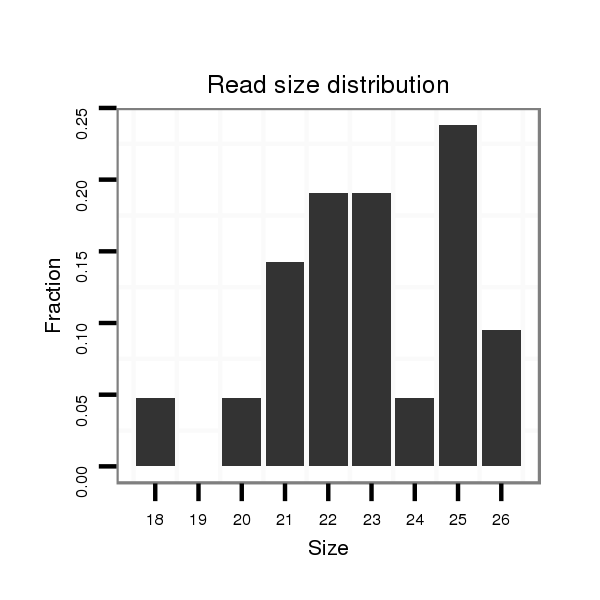
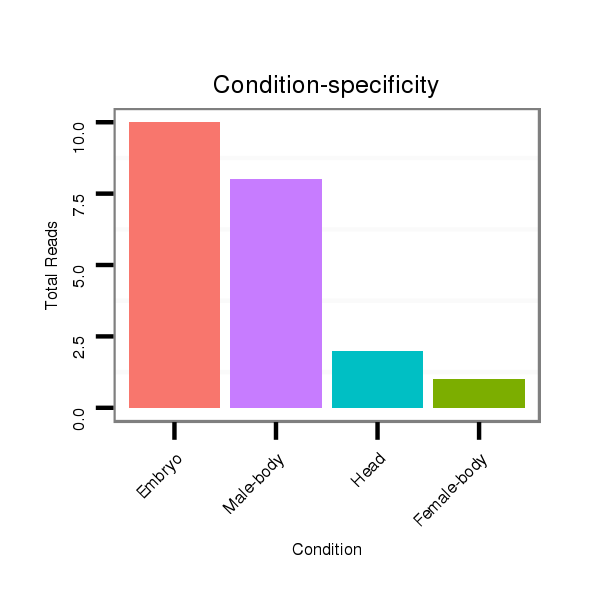
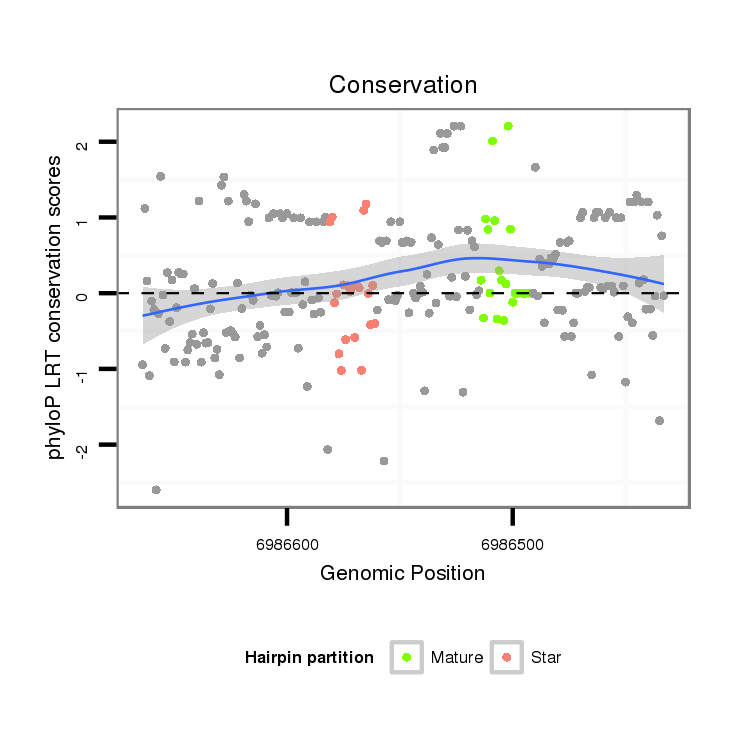
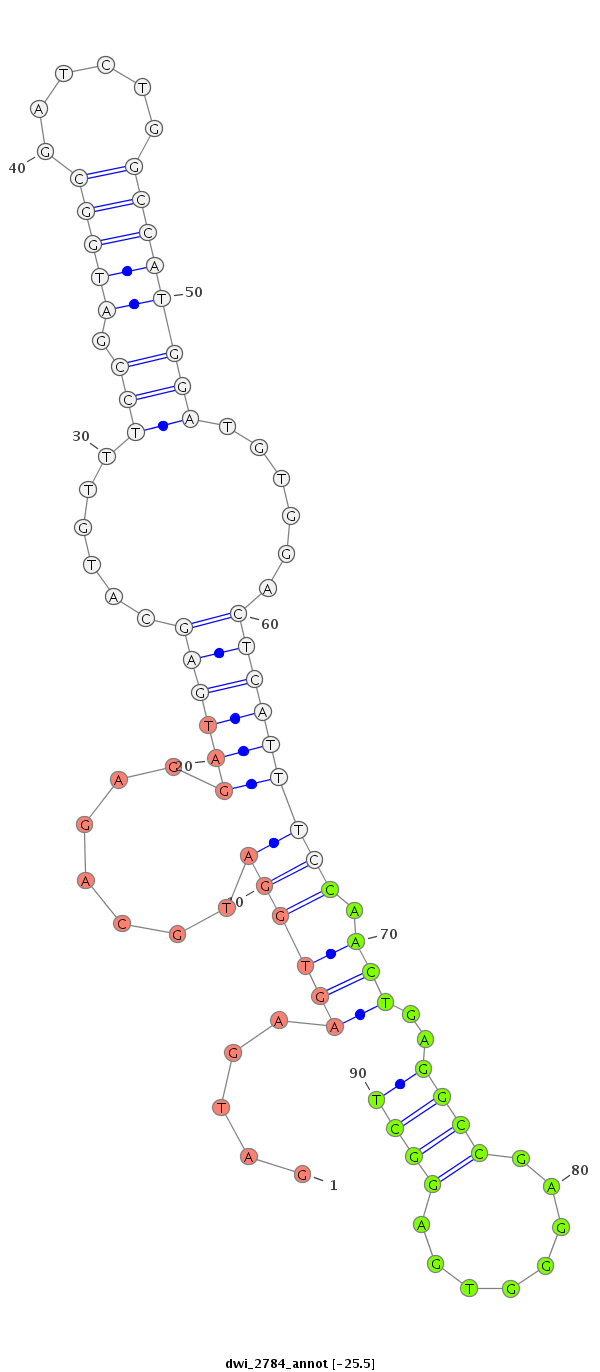
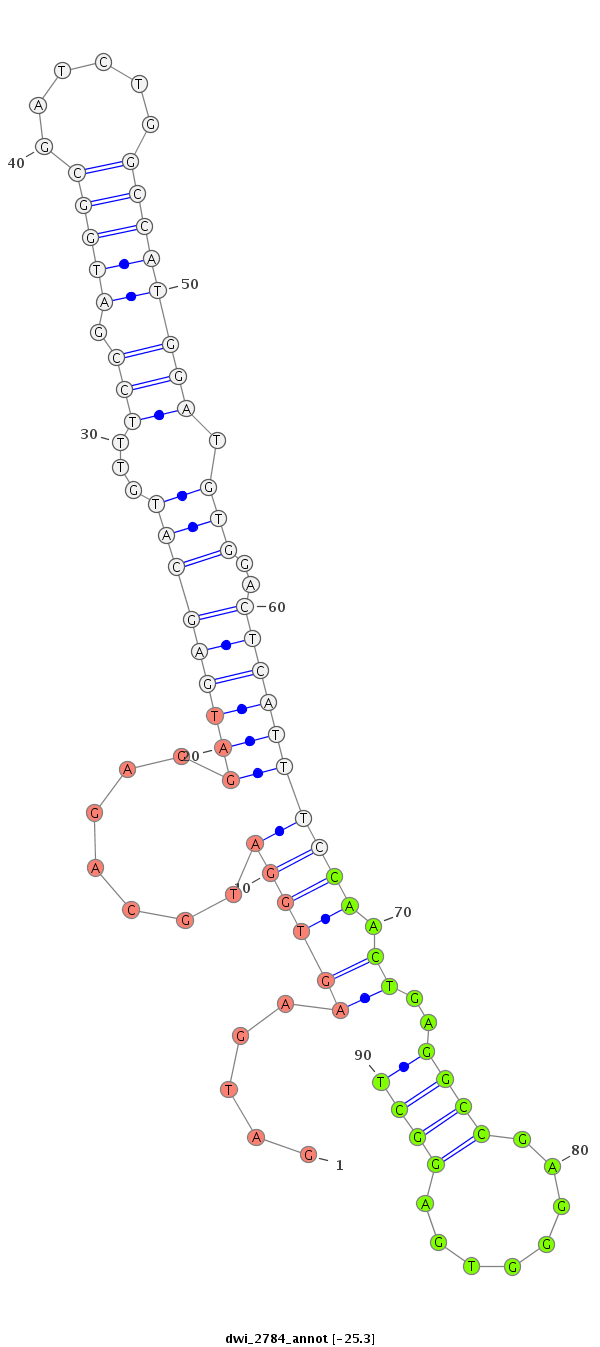
| ....AGACGAAGAGGATGACGATCT............................................................................................................................................................................................................... |
21 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................CAACTGAGGCCGAGGGTGAGGCT........................................................... |
23 |
0 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
| ...................................................................................GATGAAGTGGATGCAGAGGAT................................................................................................................................ |
21 |
0 |
1 |
2.00 |
2 |
1 |
1 |
0 |
0 |
0 |
| ...................................................................................GATGAAGTGGATGCAGAGGATGAGC............................................................................................................................ |
25 |
0 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
| ............................................................................................................ATGTTTCCGATGGCGATCTGGC...................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| .................................................................................................AGAGGATGAGCATGTTTCCGATGG............................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................................GAAGGGGATGATGAAGTGGATGC....................................................................................................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................TCAAAGAAGGGGATGATGAAGTG............................................................................................................................................ |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................CAGGAGGGGGTAATCGAATGTGA........................................................................................................................................................................ |
23 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..........................................AGGAGGGGGTAATCGAATG........................................................................................................................................................................... |
19 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| ................................................................................................................................................................CGAGGGTGAGGCTGAGTCCGAGGGC............................................... |
25 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ................................................................................................CAGAGGATGAGCATGTTTCCGATGGC.............................................................................................................. |
26 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
| ..........AGAGGATGACGATCTGCACATGGAT..................................................................................................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................................................................GTCGAGCCTGAAATTGAAACGGAATC..................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............................................................GTGATACGGTCAAAGAAGGGG....................................................................................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................................................................................ATGAAGTGGATGCAGAGGATGA.............................................................................................................................. |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................AACTGAGGCCGAGGGTGAGG............................................................. |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................GGCCAAGGAAGGGGATGATAAAG.............................................................................................................................................. |
23 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..................................................................................TGATGAAGTGGATGCAGAGGAT................................................................................................................................ |
22 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
| ..........................................................ATGTGATACGGTCAAAGAAGGGGAT..................................................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................................................................GTCGAGCCTGAAATTGAAACGGA........................ |
23 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
| ......................................................................................GAAGTGGATGCAGAGGAT................................................................................................................................ |
18 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| .......................................................................................................................................................AACTGAGGCCGAGGGTGAGGCT........................................................... |
22 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................ATCAGCAACAGATCAGGA...... |
18 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................GTGAGGCTGAGTCCGAGGG................................................ |
19 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................CGAATGTGATACGGTCAAAGAAGGGG....................................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................CAACTGAGGCCGAGGGTGAGGCTGA......................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................................AGAGGATGAGCATGTTTCCGATGGC.............................................................................................................. |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................GAGGCTGAGTCCGAGGGCGTCGAGC........................................ |
25 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ....................................................................................................................GAAGGCGATCTGGTCATGGC................................................................................................ |
20 |
3 |
3 |
0.33 |
1 |
0 |
0 |
1 |
0 |
0 |
| ...............................................................................................................................................................................................................AATCAGCAACAGCTCGGG....... |
18 |
2 |
3 |
0.33 |
1 |
0 |
0 |
1 |
0 |
0 |
| ....................................................................................................GGCTAAGCATGTTACCGA.................................................................................................................. |
18 |
3 |
20 |
0.30 |
6 |
0 |
0 |
6 |
0 |
0 |
| ............................................................................................GATGCAGTGTATGAGCAT.......................................................................................................................... |
18 |
2 |
9 |
0.22 |
2 |
0 |
1 |
0 |
0 |
1 |
| .............................................................................GGTGATGATGAATTGGCTGCA...................................................................................................................................... |
21 |
3 |
7 |
0.14 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..................................................................................TGATGAAGTGGAAGCTGGGGA................................................................................................................................. |
21 |
3 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
0 |
| ........................................................................................................................................................................AGGCAGAGTCTGAGTGCGTC............................................ |
20 |
3 |
8 |
0.13 |
1 |
0 |
0 |
1 |
0 |
0 |
| .....................................................................TGAAAGAAGCGGATGATGA................................................................................................................................................ |
19 |
2 |
8 |
0.13 |
1 |
0 |
0 |
1 |
0 |
0 |
| .........................................................................CGAAGAGGATGATGAAGTGGA.......................................................................................................................................... |
21 |
2 |
13 |
0.08 |
1 |
0 |
0 |
1 |
0 |
0 |
| ................................................................................GAAGATGAAGTGGAAGCAGA.................................................................................................................................... |
20 |
2 |
17 |
0.06 |
1 |
0 |
1 |
0 |
0 |
0 |
| .....................AGATGCGCATGGATGATCT................................................................................................................................................................................................ |
19 |
3 |
17 |
0.06 |
1 |
0 |
1 |
0 |
0 |
0 |
| ............................................................................................................................................................................................ATGCCTGAAATTGAAGCGGA........................ |
20 |
3 |
19 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................................GGTTGATGAAGTGGAAGCTG..................................................................................................................................... |
20 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................................GAAGGGGATGTTGCGGTGGA.......................................................................................................................................... |
20 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |