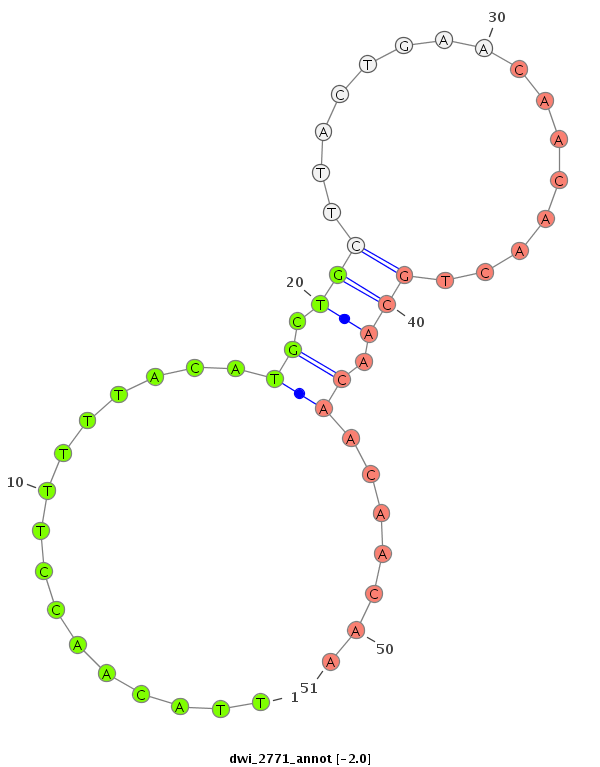
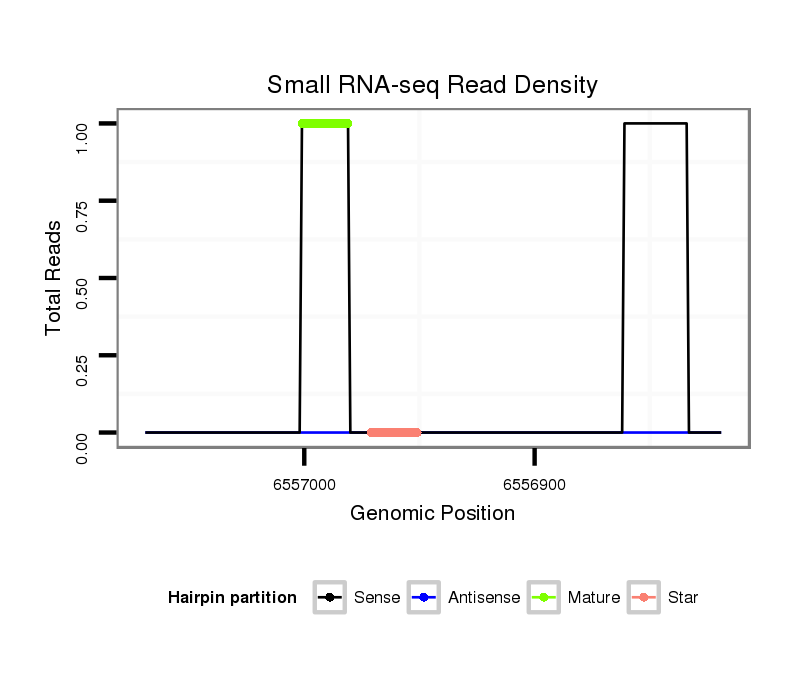
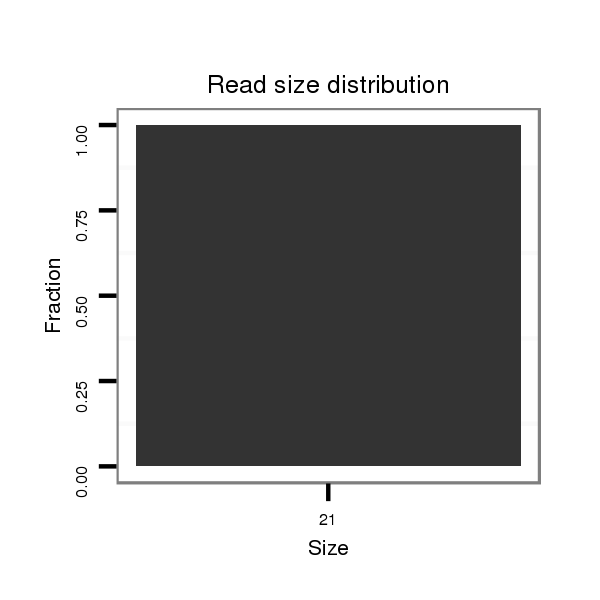
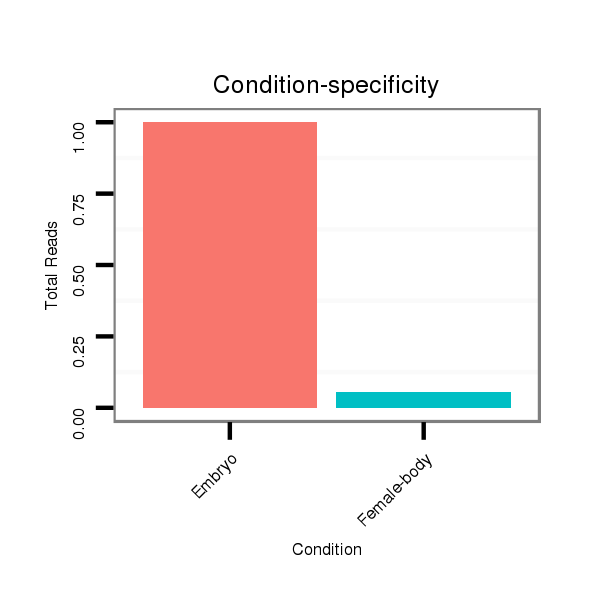
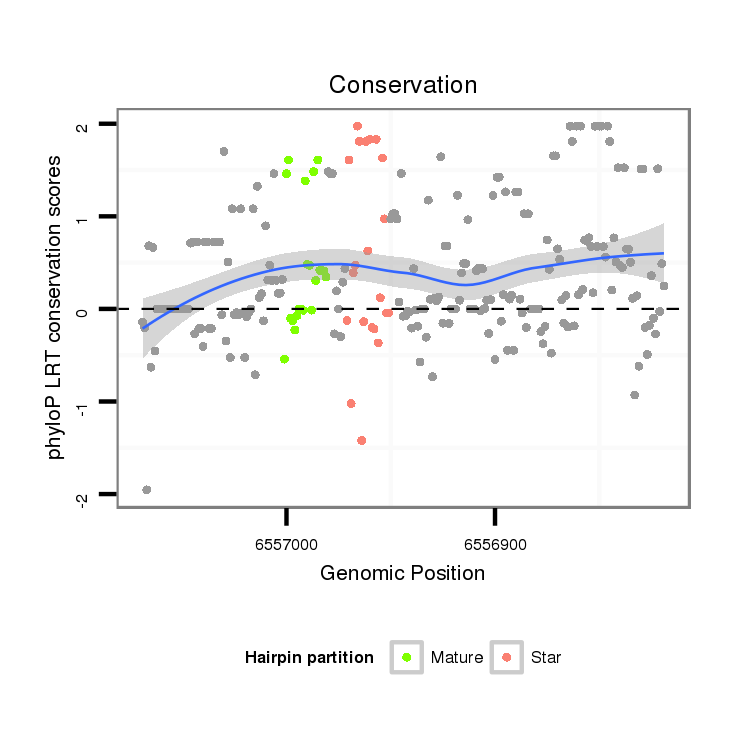
ID:dwi_2771 |
Coordinate:scf2_1100000004909:6556869-6557019 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -1.6 |
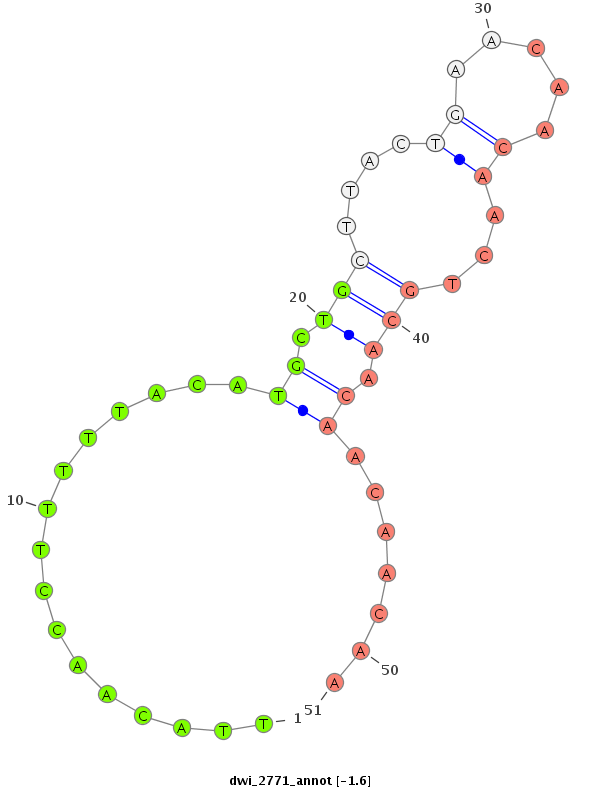 |
intron [Dwil\GK25198-in]; CDS [Dwil\GK25198-cds]; exon [dwil_GLEANR_9358:2]; intron [Dwil\GK25198-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| (TTG)n | Simple_repeat | Simple_repeat | + |
| AT_rich | Low_complexity | Low_complexity | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------##############################-------------------- GGGTGGGTGGTAAAGTGCCTTTTGTTTCCTTACATATATAAAATTATATACATAATATATTTCTTCTTTTACAACCTTTTTACATGCTGCTTACTGAACAACAACTGCAACAACAACAACAACTATGTATAATAATAATAATGACAACCTGATCTTCTCCTTACCTTTAAAATAATAATAATATTAAATTGTCTTATAAAGCTCTACTAAAAAAACTACAACAACAACCAGGTATTTGGAAATATTGCTAC ********************************************************************................((.(((................))).)).......************************************************************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V118 embryo |
V119 head |
V117 male body |
|---|---|---|---|---|---|---|---|---|---|
| ....GGGTGATAGCGTGCCTTTTGT.................................................................................................................................................................................................................................. | 21 | 3 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 |
| .....GGTGATAGCGTGCCTTTTGT.................................................................................................................................................................................................................................. | 20 | 3 | 10 | 2.90 | 29 | 25 | 0 | 1 | 3 |
| ...TGGGTGATAGCGTGCCTTTTGT.................................................................................................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................AAAAAAACTACAACAACAACCAGGTATT............... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....GGTGATAGAGTGCCTTTTGT.................................................................................................................................................................................................................................. | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................TTACAACCTTTTTACATGCTG.................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......GTTGGAAAGTGCCTTTTGTT................................................................................................................................................................................................................................. | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............AATGCCTTTTGTTTTCTTACA......................................................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........TGGAAAGTGCCTTTTGTTCCC.............................................................................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ......GTTGGAAAGTGCCTTTTGT.................................................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ......GTGATAACGTGCCTTTTGT.................................................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...TGGGTGATAGCGTGCCTTTTG................................................................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ....GGGTGATAGCGTGCCTTTTG................................................................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................ATTTTCTCCTGACCTTGAAAAT.............................................................................. | 22 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| .......TTGGAAAGTGCCTTTTGTTCC............................................................................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| .......TTGGAAAGTGCCTTTTGTT................................................................................................................................................................................................................................. | 19 | 2 | 16 | 0.13 | 2 | 1 | 1 | 0 | 0 |
| ....GGGTGATAGCGTGCCTTTT.................................................................................................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................CAACAACTGCAACAACAACAA.................................................................................................................................... | 21 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ......GTGATACCGTGCCTTTTGT.................................................................................................................................................................................................................................. | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ........TGGAAAGTGCCTTTTGTT................................................................................................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
CCCACCCACCATTTCACGGAAAACAAAGGAATGTATATATTTTAATATATGTATTATATAAAGAAGAAAATGTTGGAAAAATGTACGACGAATGACTTGTTGTTGACGTTGTTGTTGTTGTTGATACATATTATTATTATTACTGTTGGACTAGAAGAGGAATGGAAATTTTATTATTATTATAATTTAACAGAATATTTCGAGATGATTTTTTTGATGTTGTTGTTGGTCCATAAACCTTTATAACGATG
************************************************************************************************************************************................((.(((................))).)).......******************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V117 male body |
V118 embryo |
M045 female body |
|---|---|---|---|---|---|---|---|---|
| .....................................................................................CGACGAAGGACTGGTTGATG.................................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ................................................................................................TTGTTGTTGACGTTGTTGTT....................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ............................................................................................................TTGTTGTTGTTGTTGATA............................................................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| .................................................................................................TGTTGTTGACGTTGTTGTT....................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004909:6556819-6557069 - | dwi_2771 | GGGTGGGTGGTAAAGTGCCTTTTGTTTCCTTACATATATAAAATTAT----ATACATA---A----TAT----ATTTCTTCTTTTACAACCTTTTTACATGCTGCTTA-----CTGAACAACAACTGCAACAACAACAACAACTATG---------------------TATAATAATAATAATGACAACCTGAT---CTTCTCCTTACC--TTTAAAAT-------------------------------------AATAATAATATTAAATTGTCTTATAAAGCTCTACTAAAAAAACTACAACAACAACCAGGT---------ATTTGGAAATATTGCTAC |
| droVir3 | scaffold_13042:3606470-3606635 + | ATT-----------------------------------T----------------------------------CTTTCTTCCTGTACAAC--CTTTACATGCTGCTTTGGAAAA-TAAAATCAACCGCAATTAAAA-------TACATACATACATATATATA-TATCTCTTTAT--TACAATAAC--------AACTTT----CCTCTT--------------------------------------------------------------------GAAAAGCTCTACTAAAAAAACAACAACTACAACCAGGT---------ATTA----TGC--TCTAT | |
| droMoj3 | scaffold_6308:831099-831297 + | ATA-----------------------------------TATATTAAAACATATTTA-AATCTTTTCTCTATTTCTTTCTT--CCTATTAC--CTATACATGCTGCTTGGGAA-A-TAAAAAAAACTGCTATTAAAA-------TACATATATATCTATAATTATTATCT--AT-----ACTATAAT--------AACTTT----CCTCAC--------------------GAA---------------------------------------------AAAAAGCTCTACTAAAAAAACAACAACTACAACCAGGT---------ATTC--AATGCTCTCTAC | |
| droGri2 | scaffold_15203:1685428-1685492 + | TTGTCGT---------------------------------------------------------------------AC-------ACATT--TTTCACATATTCC------------ACAGCGACAAC------------------------------------------------------------------------------------------------------------------------------------------------------------GACAACAACAGCAACAACAACAACAAC-------------------------------- | |
| droPer2 | scaffold_820:4829-4975 - | AAC-----------------------------------AACA---------------------------------------------------------------------------------ACAACAACAACAACAACAACAACAAGTAGACCAACAACAATCATCACA-------ACAACAACAAC---AC---CACC---TCCTCCTCCAACAAC-------------------------------------AACAACAACATCA-------------TTCCACACAACATAATCCACAACAACAG-----C---------ATCTGCAACAGCAGCAGC | |
| droBip1 | scf7180000395529:233943-234116 - | ATA-----------------------------------AATA-------------------ACTTTTCCCTCT--ATTTCTCTCTATTCT--C-TTCCCTGCTGCTTGGAATAC-GATAATCAACCGCAATTATT---------------------------------TCAATAC-----TATAAT--A---AT---CTTC---TCTCCTATTTAAAAA-------------------------------------AAAATAAAT-------------ATAAAGCTCTACTAAAAAAACAACAACAACAACCAGGT---------ACAAAAAAAAATAACTA- | |
| droKik1 | scf7180000302696:443207-443430 + | TTT--------------------GTATTTGTATGTATATACCCTTTTACCTAACTT-AATACTCTCTCTCTCT--TTTTTCCTCTATTCT--CTTTTCCTGCCGCTTGGAATAC-TTAAAACAACCGCAATTATTA---------------------------------TAATAT-----CTTAAT--A---ATCTCCTT----TCTCCTATCTAAAAACAATGCAATATAAA---------------------------------------------AAAAAGCTCTACTAAAAAAACAACAACAACGACCAGGTACAAACAACG-TAGAAAATATAACAAC | |
| droBia1 | scf7180000302378:208407-208548 + | AAC-----------------------------------AACA---------------------------------------------------------------------------------ACAACAACAACAACGGCAACAATG---------------------CCGAATC-----TGGT-------------------------------GAAACTAAGCAGTAAAAAGCAAATTCCTGTTAATAAATTACTACCACAATGTTA-----AACTAGAAAGAATGACAAAAAACATAATAATAACTAACCAGT---------ATTT-------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
Generated: 05/18/2015 at 05:19 AM