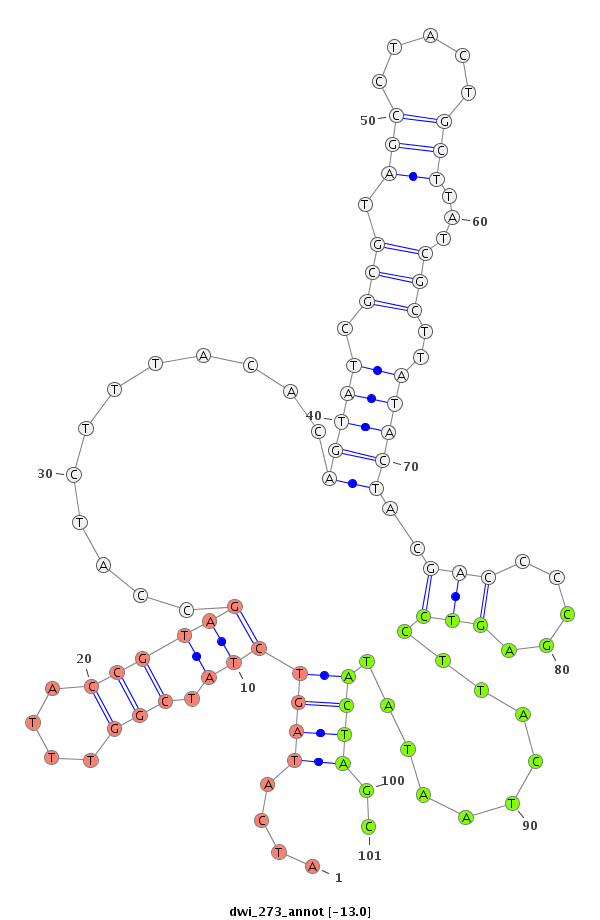
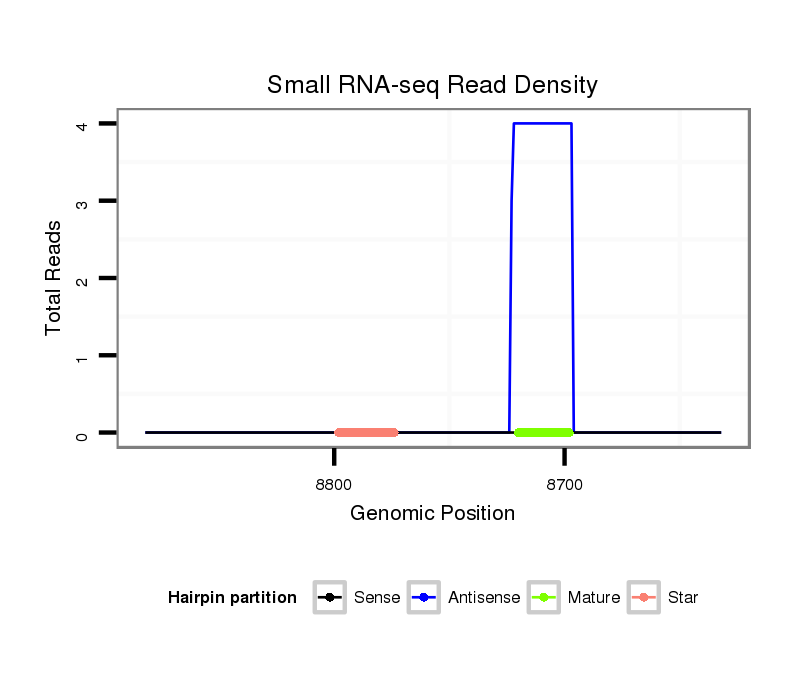
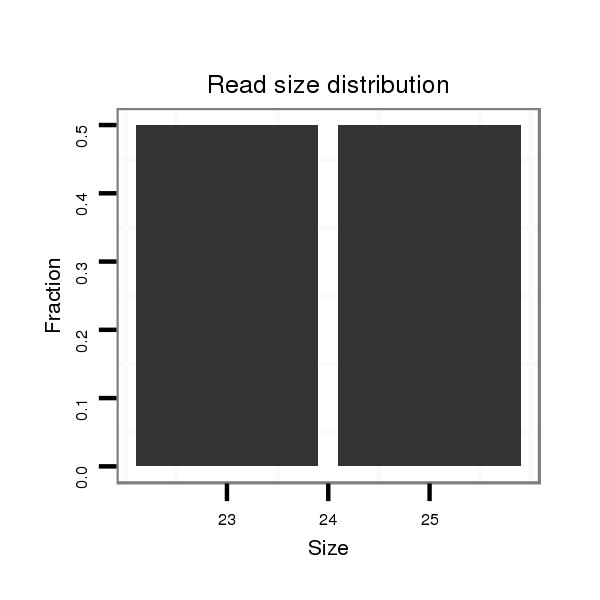
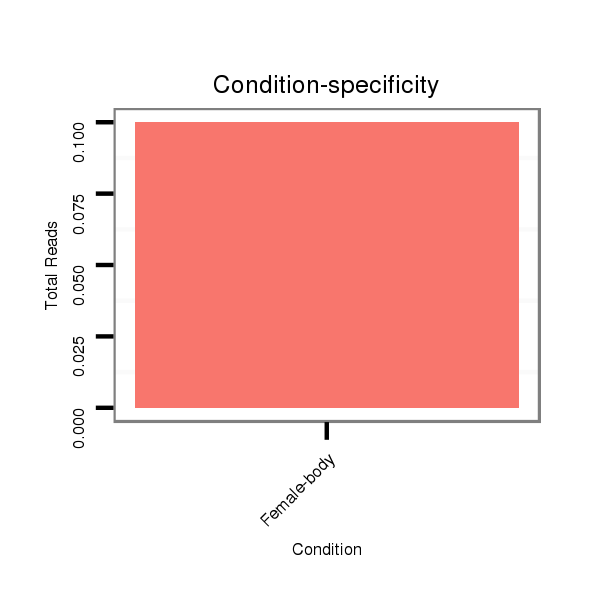
ID:dwi_273 |
Coordinate:scf2_1100000002710:8682-8832 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -12.9 | -12.3 | -12.3 |
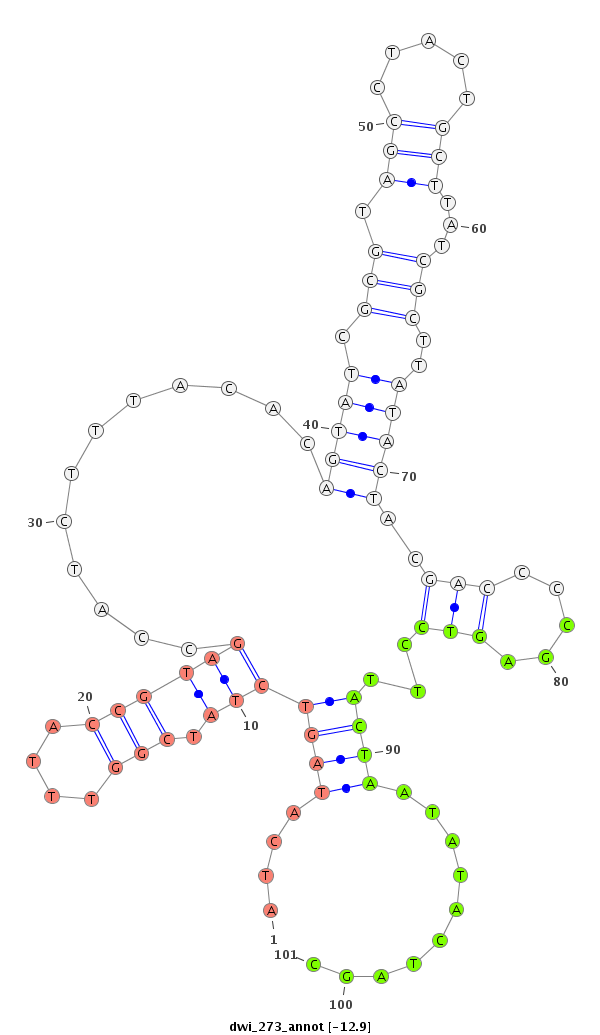 |
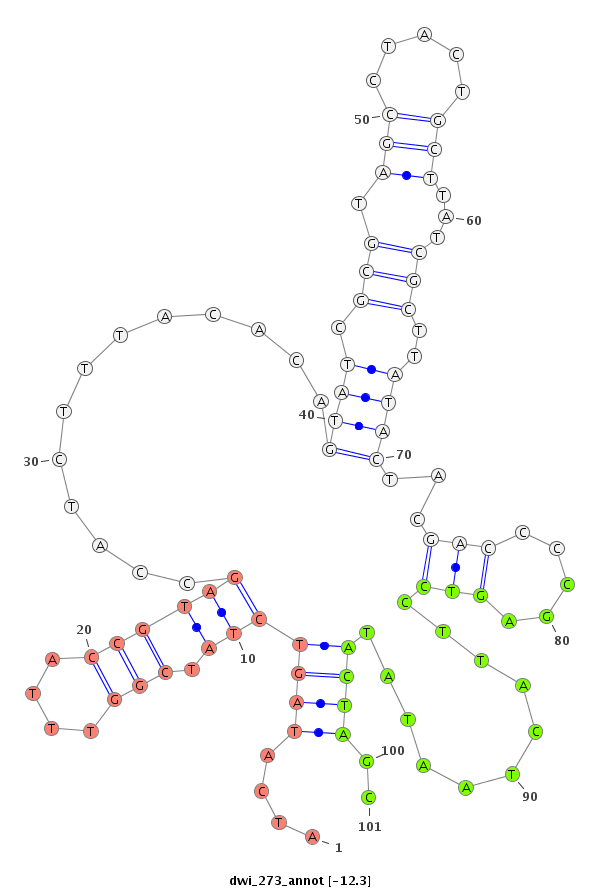 |
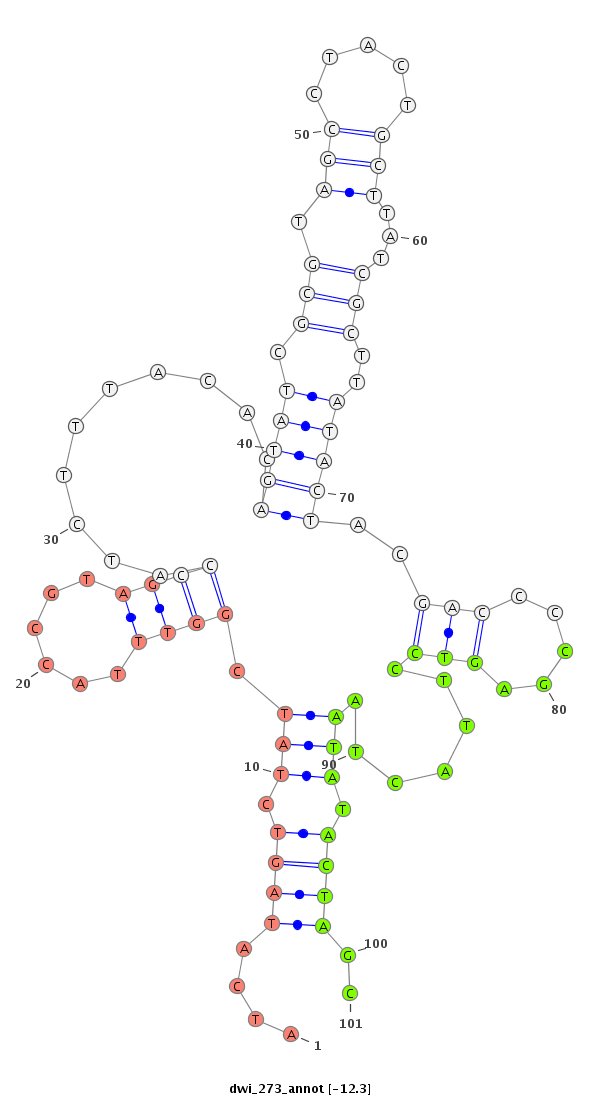 |
CDS [Dwil\GK14336-cds]; exon [dwil_GLEANR_1466:2]; intron [Dwil\GK14336-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTTATATTTTCTGACCCCTCCTTTGCAGAATTTATTAAAAATGAAATAAAATATATATAGGCATTGGTCAAATTGCACATCAATATCATAGTCTATCGGTTTACCGTAGCCATCTTTACACAGTATCGCGTAGCCTACTGCTTATCGCTTATACTACGACCCCGAGTCCTTACTAATATACTAGCAATGGGCGTTCATTAGATAAACCATCGTCGCAAAGCCTTTGTGACGAATTCGGATTAGACACCGGT ************************************************************************************....(((((((.(((....))))))............(((((.(((.(((.....)))...)))..)))))..(((.....)))...........))))..****************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
M020 head |
M045 female body |
|---|---|---|---|---|---|---|---|---|
| .............................................................................................................CCATCGTGACACTGTATCGC.......................................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................TGTGACGAATTCGGATTAGACACCGG. | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| ..................................................................................................................................................................CGAGTCCTTACTAATATACTAGC.................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................................TGTGACGAATTCGGATTAGACGCCGG. | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................................TGTGACGAATTCGGATTAGAAACCGG. | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ....................................................................................ATCATAGTCTATCGGTTTACCGTAG.............................................................................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ...............................................................................................................................................................................................................................TTGTGACGAATTCGGATTAGACACCGG. | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
|
AAATATAAAAGACTGGGGAGGAAACGTCTTAAATAATTTTTACTTTATTTTATATATATCCGTAACCAGTTTAACGTGTAGTTATAGTATCAGATAGCCAAATGGCATCGGTAGAAATGTGTCATAGCGCATCGGATGACGAATAGCGAATATGATGCTGGGGCTCAGGAATGATTATATGATCGTTACCCGCAAGTAATCTATTTGGTAGCAGCGTTTCGGAAACACTGCTTAAGCCTAATCTGTGGCCA
******************************************************************....(((((((.(((....))))))............(((((.(((.(((.....)))...)))..)))))..(((.....)))...........))))..************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V118 embryo |
|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................GGGGCTCAGGAATGATTATATGATCGT................................................................. | 27 | 0 | 18 | 3.67 | 66 | 56 | 10 |
| ................................................................................................................................................................GGGCTCAGGAATGATTATATGATCGT................................................................. | 26 | 0 | 19 | 1.32 | 25 | 16 | 9 |
| ............................................................................................GATAGCCAAATGGCATCGGTAGAAAT..................................................................................................................................... | 26 | 0 | 20 | 0.85 | 17 | 6 | 11 |
| ...............................................................................................AGCCAAATGGCATCGGTAGAAATGTGT................................................................................................................................. | 27 | 0 | 8 | 0.75 | 6 | 2 | 4 |
| ............................................................................................................................................................................................................................GGAAACACTGCTTAAGCCTAATCTGT..... | 26 | 0 | 20 | 0.50 | 10 | 6 | 4 |
| .........GGACTGGGAACGAAACGTCTT............................................................................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 |
| ...........................................................................................AGATAGCCAAATGGCATCGGTAGAAAT..................................................................................................................................... | 27 | 0 | 20 | 0.45 | 9 | 3 | 6 |
| .................................................................................................................................................................GGCTCAGGAATGATTATATGATCGT................................................................. | 25 | 0 | 19 | 0.37 | 7 | 5 | 2 |
| ..........................................................................................CAGATAGCCAAATGGCATCGGTAGAAAT..................................................................................................................................... | 28 | 0 | 19 | 0.26 | 5 | 4 | 1 |
| ...............................................................................................................................................................GGGGCTCAGGAATGATTATATGATCG.................................................................. | 26 | 0 | 19 | 0.21 | 4 | 4 | 0 |
| ..........................................................................................................................................................ATGCTGGGGCTCAGGAATGATTATAT....................................................................... | 26 | 0 | 20 | 0.20 | 4 | 0 | 4 |
| ............................................................................................................................................................................GATTATATGATCGTTACCCGCAAGT...................................................... | 25 | 0 | 14 | 0.14 | 2 | 0 | 2 |
| ...............................................................................................AGCCAAATGGCATCGGTAGAAATGT................................................................................................................................... | 25 | 0 | 14 | 0.14 | 2 | 2 | 0 |
| ................................................................................................GCCAAATGGCATCGGTAGAAATGTGT................................................................................................................................. | 26 | 0 | 8 | 0.13 | 1 | 0 | 1 |
| ..........................................................................................................................CGTAGAGAATCGGATGACG.............................................................................................................. | 19 | 3 | 18 | 0.11 | 2 | 2 | 0 |
| ................................................................................................................................................................................................CAAGTAAACTATTTGGTAGCAGCGTT................................. | 26 | 1 | 14 | 0.07 | 1 | 0 | 1 |
| ...........................................................................................................................................CGAATAGCGAATATGATGCTGGGGC....................................................................................... | 25 | 0 | 15 | 0.07 | 1 | 0 | 1 |
| .........................................................................................................................................................GATGCTGGGGCTCAGGAATGATTAT......................................................................... | 25 | 0 | 16 | 0.06 | 1 | 0 | 1 |
| .................................................................................................................................................GCGAATATGATGCTGGGGCTCAGGAAT............................................................................... | 27 | 0 | 17 | 0.06 | 1 | 0 | 1 |
| ..............................................................................................................................................................TGGGGCTCAGGAATGATTATATGATCGT................................................................. | 28 | 0 | 18 | 0.06 | 1 | 1 | 0 |
| ...............................................................................................................................................................GGGGCTCAGGAATGACTATATGATCGT................................................................. | 27 | 1 | 18 | 0.06 | 1 | 1 | 0 |
| .......................................................................................................................................................ATGATGCTGGGGCTCAGGAATGAT............................................................................ | 24 | 0 | 18 | 0.06 | 1 | 1 | 0 |
| ...............................................................................................................................................................GGGGCTCAGGAATTATTATATGATCGT................................................................. | 27 | 1 | 18 | 0.06 | 1 | 1 | 0 |
| ....................................................................................................................................................AATATGATGCTGGGGCTCAGGAATGA............................................................................. | 26 | 0 | 18 | 0.06 | 1 | 0 | 1 |
| ..................................................................................................................................................................GCTCAGGAATGATTATATGATCGT................................................................. | 24 | 0 | 19 | 0.05 | 1 | 1 | 0 |
| ................................................................................................................................................................TGGCTCAGGAATGATTATATGATCGT................................................................. | 26 | 1 | 19 | 0.05 | 1 | 1 | 0 |
| ........................................................................................................................................................................GAATGATTATATGATCGTTACCCGC.......................................................... | 25 | 0 | 19 | 0.05 | 1 | 1 | 0 |
| ...............................................................................................................................................................GGGGCTCAGGAATGATTATATTATCGT................................................................. | 27 | 1 | 19 | 0.05 | 1 | 1 | 0 |
| .....................................................................................................................................................ATATGATGCTGGGGCTCAGGAAT............................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| ...........................................................................................................................................................TGCTGGGGCTCAGGAATGATTATAT....................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| .................................................................................................................................................................GGCTCAGGAATGATTATATGATCG.................................................................. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| ..........................................................................................................................................................ATGCTGGGGCTCATGAATGATTATAT....................................................................... | 26 | 1 | 20 | 0.05 | 1 | 0 | 1 |
| .............................................................................................ATAGCCAAATGGCATCGGTAGAAA...................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| ...........................................................................................AGATAGCCAAATGGCGTCGGTAGAAAT..................................................................................................................................... | 27 | 1 | 20 | 0.05 | 1 | 0 | 1 |
| ..........................................................................................................................................................ATGCTGGGGCTCAGGAATGAT............................................................................ | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| ............................................................................................................................................................................................................................GGAAACACTGCTTAAGCCTAATCTG...... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| ..........................................................................................................................................................ATGCTGGGGCTCAGGAATGATTAT......................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| ............................................................................................................................................................GCTGGGGCTCAGGAATGATTATAT....................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| ..............................................................................................................................................................TGGGGCTCAGGAATGATTATATGAT.................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| ...............................................................................................................................................................GGGGCTCAGGAATGATTATATGAT.................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000002710:8632-8882 - | dwi_273 | TTTATATTTTCTGACCCCTCCTTTGCAGAATTTATTAAAAATGAAATAAAATATATATAGGCATTGGTCAAATTGCACATCAATATCATAGTCTATCGGTTTACCGTAGCCATCTTTACACAGTATCGCGTAGCCTACTGCTTATCGCTTATACTACGACCCCGAGTCCTTACTAATATACTAGCAATGGGCGTTCATTAGATAAACCATCGTCGCAAAGCCTTTGTGACGAATTCGGATTAGACACCGGT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 05/17/2015 at 10:33 PM