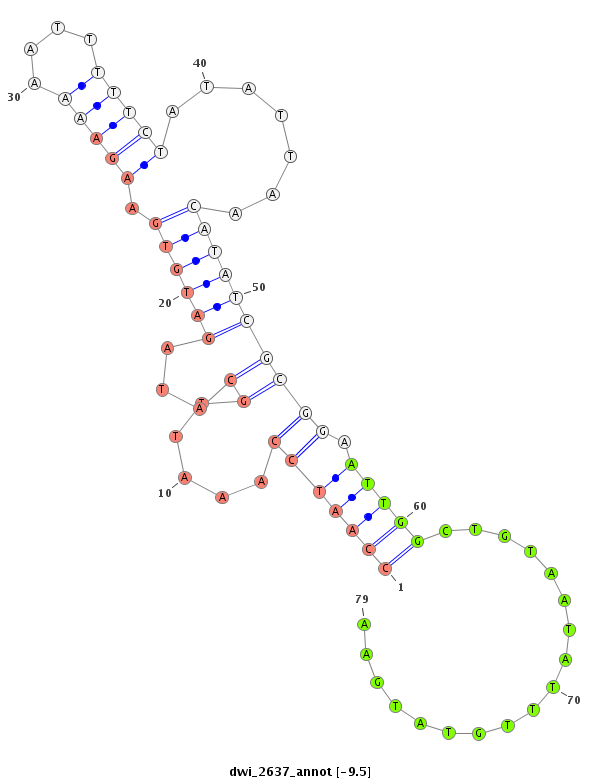
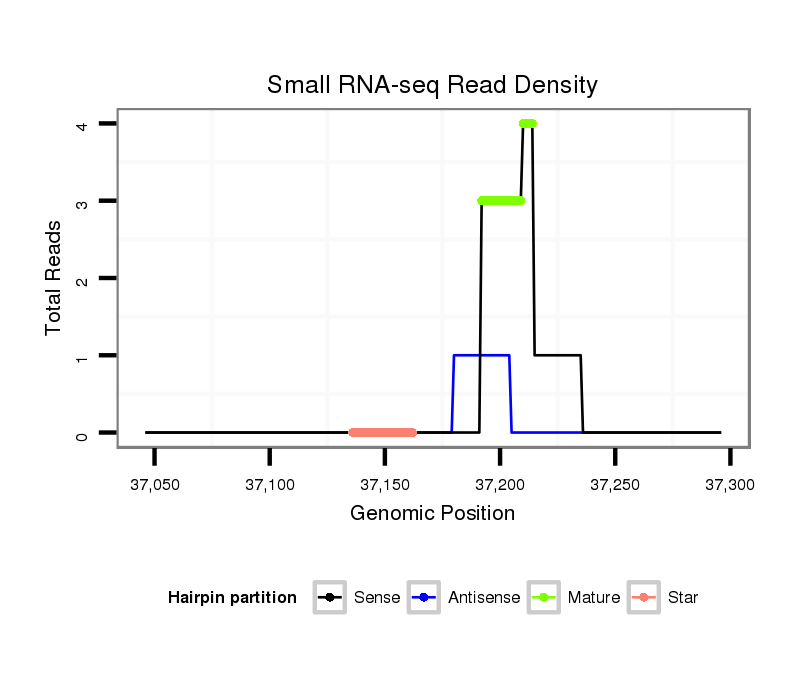
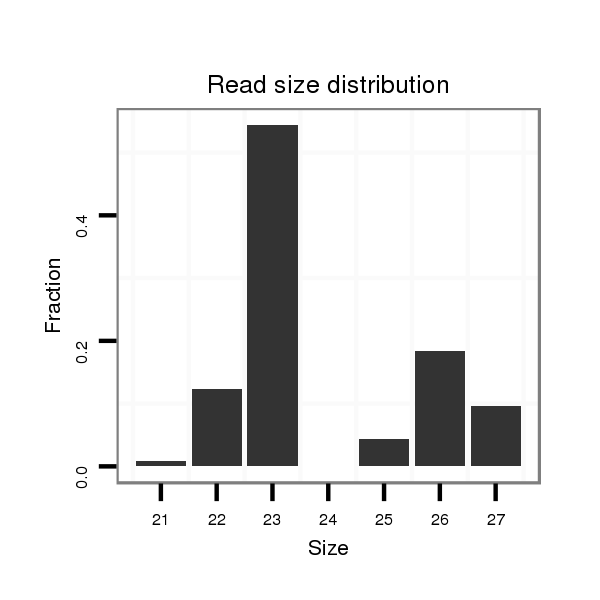
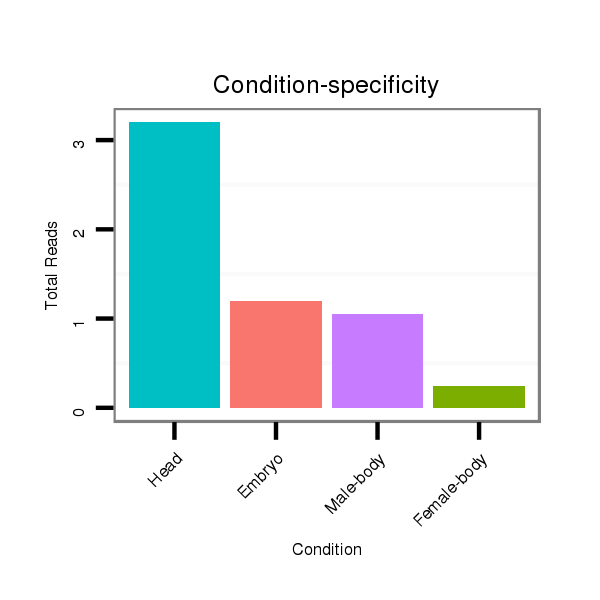
ID:dwi_2637 |
Coordinate:scf2_1100000004908:37096-37246 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -9.5 | -9.4 | -9.4 |
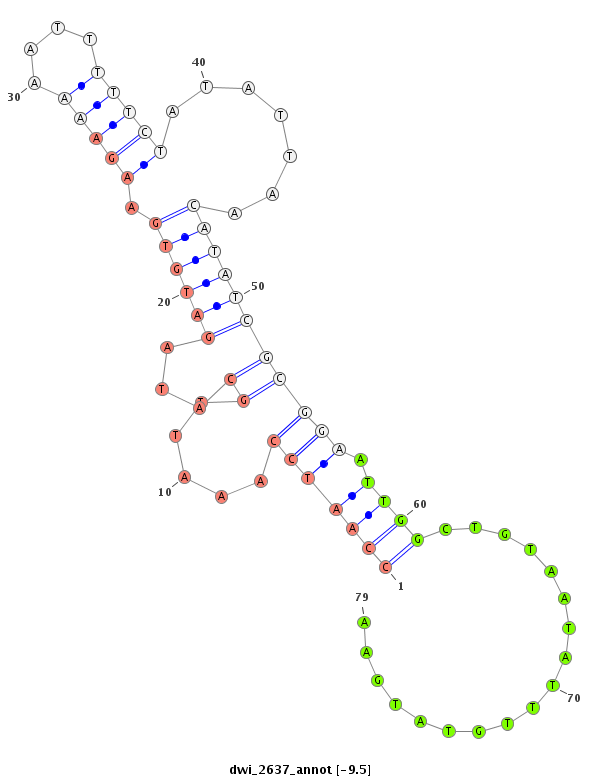 |
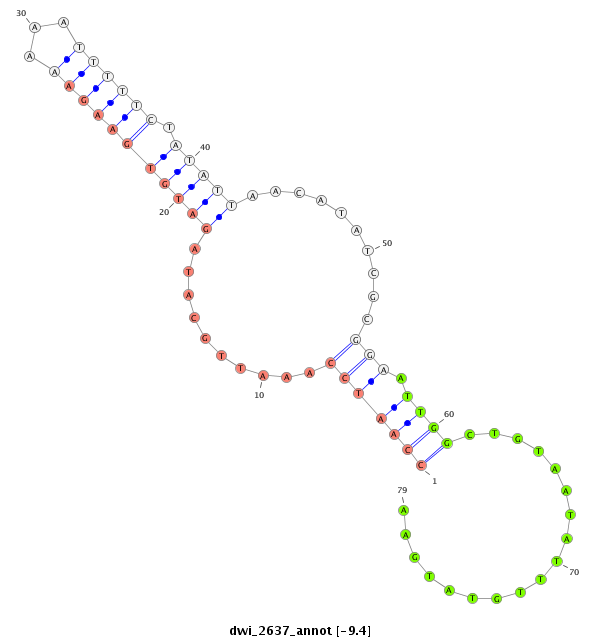 |
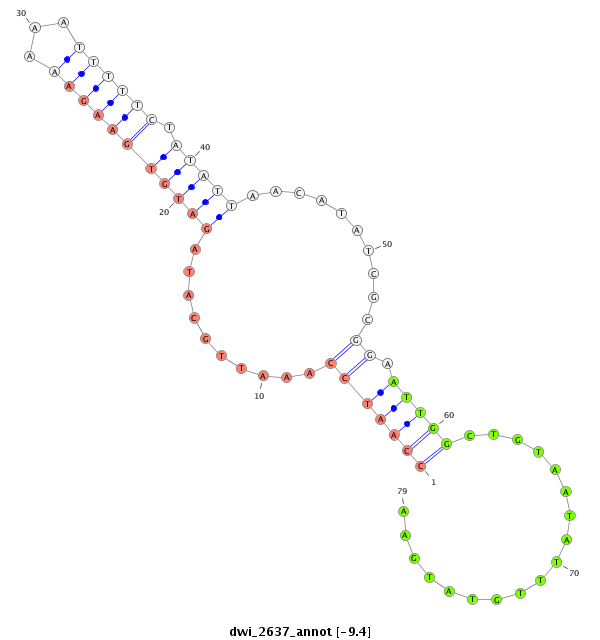 |
intron [Dwil\GK18732-in]; exon [dwil_GLEANR_1936:2]; CDS [Dwil\GK18732-cds]; intron [Dwil\GK18732-in]
No Repeatable elements found
| mature | star |
| ------------######################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TTCATATTTCAGATGCACATTCCACGTCGATTCTATTCGAACTTTCAACAGTAATTTCGTAAATTAATATTATAACAGACGTTTCCATTACCAATCCAAATTGCATAGATGTGAAGAAAAATTTTTCTATATTAACATATCGCGGAATTGGCTGTAATATTTGTATGAATTTAACATGGAAGAAGACTTTTCATTCAAATCCTCATTCAAAAGTGGACTGAAAATGTTGCAAAGCATCTTTTCCATAATTC ******************************************************************************************(((((((.....((...((((((.(((((....))))).......)))))))))).)))))..................********************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M020 head |
V118 embryo |
M045 female body |
V117 male body |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................ATTGGCTGTAATATTTGTATGAA.................................................................................. | 23 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ............................................................................................................AAGTGAAGAAAAATTTTT............................................................................................................................. | 18 | 1 | 13 | 1.31 | 17 | 0 | 17 | 0 | 0 |
| ....................................................................................................................................................................ATGAATTTAACATGGAAGAAGACTTT............................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ...ATATTTCAGGTGCACATTCCACGTCGA............................................................................................................................................................................................................................. | 27 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................CCAATCCAAATTGCATAGATGTGAAGA...................................................................................................................................... | 27 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................TTGCATAGATGTGAAGAAAAAT................................................................................................................................. | 22 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ...................TTCCACGTCGATTCTATTCGAACT................................................................................................................................................................................................................ | 24 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................................................................TATTAACATATCGCGGAATTGGCTG................................................................................................. | 25 | 0 | 20 | 0.15 | 3 | 0 | 1 | 2 | 0 |
| ........................................................................................................................................ATATCGCGGAATTGGCTGTAAT............................................................................................. | 22 | 0 | 20 | 0.15 | 3 | 2 | 0 | 0 | 1 |
| .................................................................................................................................TATTAACATATCGCGGAATTGGCTC................................................................................................. | 25 | 1 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| .................................................................................................................................TATTAACATATCGCGGAATTGGC................................................................................................... | 23 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 |
| ............................................................................................................ATGTGAAGAAAATTTTTT............................................................................................................................. | 18 | 1 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................TTACATGGTAGAAGACTTT............................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................AACATATCGCGGAATTGGCTG................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................TAACATATCGCGGAATTGGCTGTAATA............................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................CCAGATGGCGGAATTGGCT.................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ............................................................................................................AAGTGAAGAAAAATTTTTTT........................................................................................................................... | 20 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................................................................ATATTAACATATCGCGGAATTG..................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................TTTCTATATTAACATATCGCGGAAT....................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................TTCTATATTAACATATCGCGGAATT...................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................TATTAACATATCGCGGAATTGGCTGT................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
AAGTATAAAGTCTACGTGTAAGGTGCAGCTAAGATAAGCTTGAAAGTTGTCATTAAAGCATTTAATTATAATATTGTCTGCAAAGGTAATGGTTAGGTTTAACGTATCTACACTTCTTTTTAAAAAGATATAATTGTATAGCGCCTTAACCGACATTATAAACATACTTAAATTGTACCTTCTTCTGAAAAGTAAGTTTAGGAGTAAGTTTTCACCTGACTTTTACAACGTTTCGTAGAAAAGGTATTAAG
**********************************************************************************(((((((.....((...((((((.(((((....))))).......)))))))))).)))))..................****************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V118 embryo |
|---|---|---|---|---|---|---|---|
| ......................................................................................................................................TGTATAGCGCCTTAACCGACATTAT............................................................................................ | 25 | 0 | 20 | 1.50 | 30 | 17 | 13 |
| .................GTAAGGTGCAGCTAAGATAAGCTT.................................................................................................................................................................................................................. | 24 | 0 | 2 | 0.50 | 1 | 0 | 1 |
| ..........................................AAAGTGGTCATTAACGCATAT............................................................................................................................................................................................ | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 |
| .................................................................................................................................................................................................AAGTTTAGGTGGAAGTTTT....................................... | 19 | 2 | 15 | 0.20 | 3 | 1 | 2 |
| ...........................................................................GTCTGCAAAGGTAATGGTTA............................................................................................................................................................ | 20 | 0 | 8 | 0.13 | 1 | 1 | 0 |
| ....ATAAAGTCTACGTGTAAGGTGCAG............................................................................................................................................................................................................................... | 24 | 0 | 8 | 0.13 | 1 | 1 | 0 |
| ..................................................................................................................................................................................................AGTTTAGGAGGAAGTTTTTA..................................... | 20 | 2 | 11 | 0.09 | 1 | 0 | 1 |
| .................................................................................................................................................................................CCTTCGTCAGAAAAGTAGG....................................................... | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 |
| ....................................AGCTTGAAATTTGGCATT..................................................................................................................................................................................................... | 18 | 2 | 19 | 0.05 | 1 | 1 | 0 |
| ....................................AGCTTCAAATTTGTCATT..................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 |
| ....................................................................................................................................ATTGTATAGCGCCCTAACCGACATTAT............................................................................................ | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 |
| ...................................................................................................................................ATTTGTATAGCGCCTTAACCGACAT............................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 |
| .................................................................................................................................ATATTTGTATAGCGCCTTAACCAACAT............................................................................................... | 27 | 2 | 20 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004908:37046-37296 + | dwi_2637 | TTCATATTTCAGATGCACATTCCACGTCGATTCTATTCGAACTTTCAACAGTAATTTCGTAAATTAATATTATAACAGACGTTTCCATTACCAATCCAAATTGCATAGATGTGAAGAAAAATTTTTCTATATTAACATATCGCGGAATTGGCTGTAATATTTGTATGAATTTAACATGGAAGAAGACTTTTCATTCAAATCCTCATTCAAAAGTGGACTGAAAATGTTGCAAAGCATCTTTTCCATAATTC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 05/18/2015 at 05:00 AM