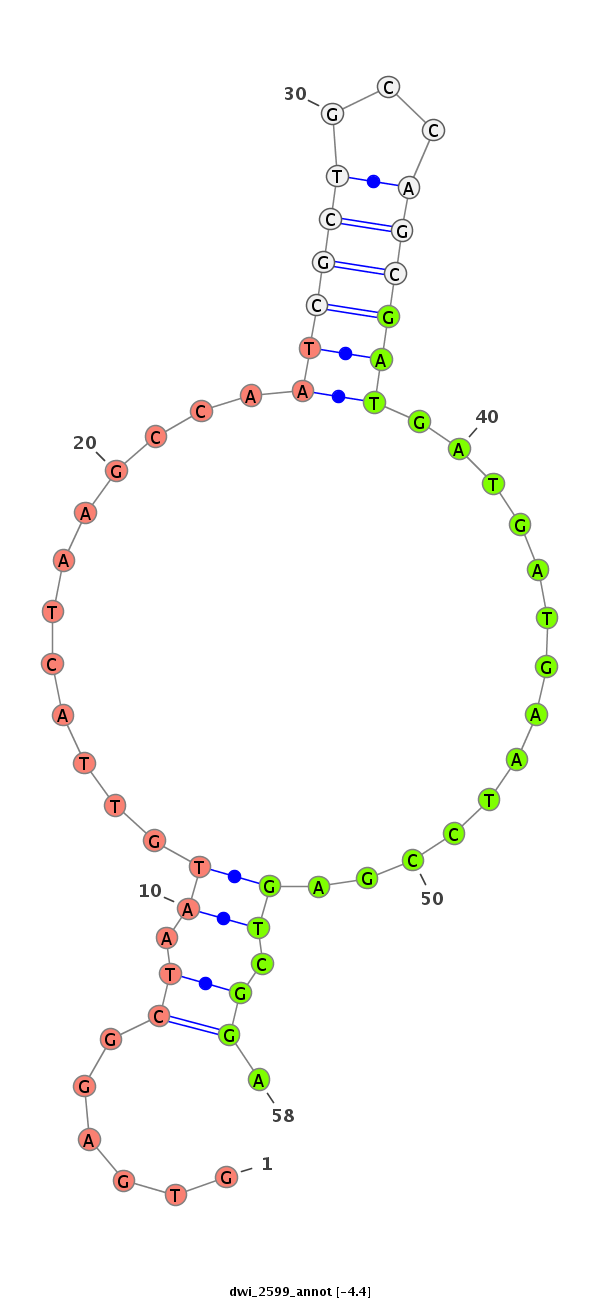
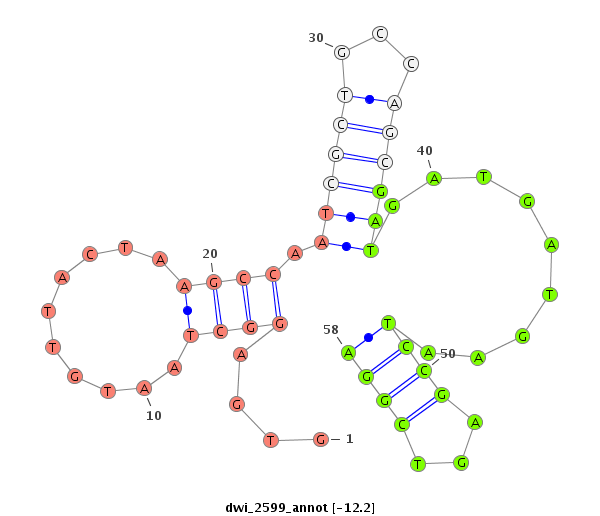
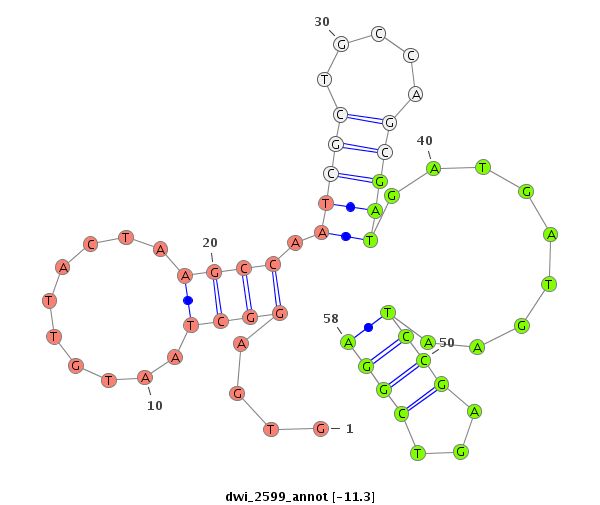
| TTCTCAA-GCTGA-GG---------------------------AGGATGATGA------T------GAGGAAGAC---------------GAGGATGATGATGATGGTG------AGGC---TAA---TGTTAC---TA---------AGCCAATCGCTGCCAGC------------------------GATGATG---------------ATGAATCC---GA---GT--------CGG---A---TGATGATGAGGACGAAGATGACGAGGAGGAGCCTGGTGAAATTAAGCCAGGC------------A---AAGTAACTC---------------AGAAA | Size | Hit Count | Total Norm | Total | M020
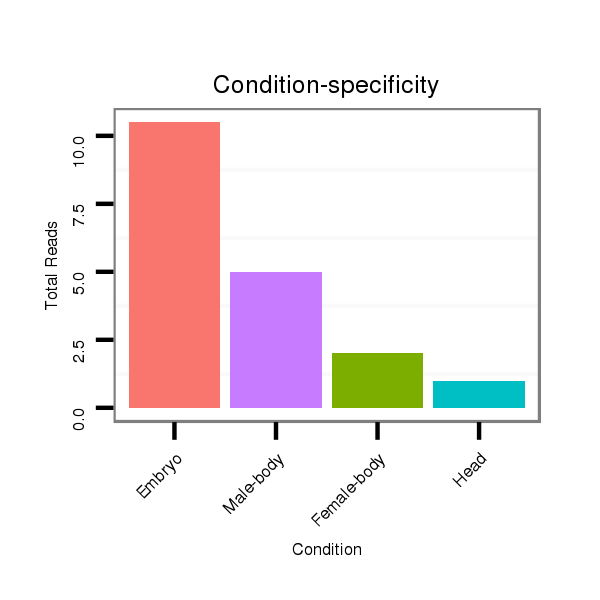
Head | M045
Female-body | V117
Male-body | V118
Embryo | V119
Head |
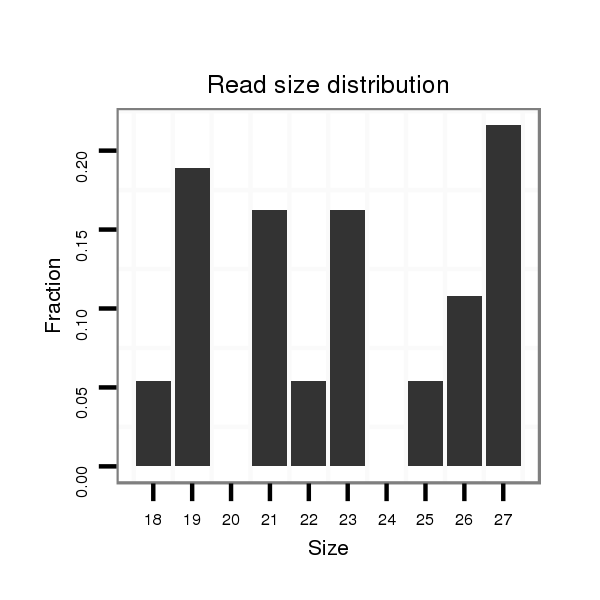
| .......................................................................AGAC---------------GAGGATGATGATGATGGTG------AG................................................................................................................................................................................................................................... | 25 | 1 | 3.00 | 3 | 0 | 0 | 1 | 1 | 1 |
| .............................................................................................................................................................................................GATGATG---------------ATGAATCC---GA---GT--------CGG---A---................................................................................................. | 23 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 |
| ......A-GCTGA-GG---------------------------AGGATGATGA------T------GAGGA................................................................................................................................................................................................................................................................................. | 24 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 |
| .............................................................................................................................................................................................GATGATG---------------ATGAATCC---GA---GT--------........................................................................................................... | 19 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 |
| ..........................................................................................................................................A---------AGCCAATCGCTGCCAGC------------------------........................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................A---TGATGATGAGGACGAAGATGAC........................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................................................GATGAGGACGAAGATGACGAGGA...................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................A---TGATGATGAGGACGAAGATGACGA......................................................................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................AGATGACGAGGAGGAGCCT............................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................ATGATG---------------ATGAATCC---GA---GT--------CGG---A---TGAT............................................................................................. | 26 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................GATGATGGTG------AGGC---TAA---T....................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................GAC---------------GAGGATGATGATGATGGT............................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................AGAC---------------GAGGATGATGATGATGGT............................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................C---------------GAGGATGATGATGATGGT............................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................TGATGAGGACGAAGATGACGAGGAGG.................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................TGCCAGC------------------------GATGATG---------------ATGAATCC---GA---GT--------........................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................GAAGATGACGAGGAGGAGCCTG.............................................................. | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................CTGCCAGC------------------------GATGATG---------------ATGAATCC---GA---GT--------........................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...TCAA-GCTGA-GG---------------------------AGGATGATGA------T------GAGGA................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................GGATGATGATGATGGTG------AG................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................TAA---TGTTAC---TA---------AGCCAATCGCTGCCAG.................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........GCTGA-GG---------------------------AGGATGATGA------T------GAGGAAG............................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................TGA------T------GAGGAAGAC---------------GAGGATGAT..................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........GA-GG---------------------------AGGATGATGA------T------GAGGAAGAC---------------GA............................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GATGATG---------------ATGAATCC---GA---GT--------CGG---A---TGAT............................................................................................. | 27 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................AGAC---------------GAGGATGATGATGATG.............................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................TAC---TA---------AGCCAATCGCTGCCAGC------------------------GATGA...................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................GATGATGGTG------AGGC---TAA---TGTT.................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................GAATCC---GA---GT--------CGG---A---TGATGATGA........................................................................................ | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................GATGA------T------GAGGAAGAC---------------GAGGATGATG.................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................GAC---------------GAGGATGATGATGATGGTG------A.................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................GATG---------------ATGAATCC---GA---GT--------CGG---A---T................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................GATGAGGACGAAGATGAC........................................................................... | 18 | 2 | 1.00 | 2 | 1 | 0 | 0 | 0 | 1 |
| ......A-GCTGA-GG---------------------------AGGATGATGA------T------GAGG.................................................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................GAGGACGAAGATGACGAGGAGG.................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................GATGA------T------GAGGAAGAC---------------GAGGATGA...................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................TGATGA------T------GAGGAAGAC---------------GAGGATGATGA................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................ATGATGATGGTG------AGGC---TA............................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................ATGACGAGGAGGAGCCTG.............................................................. | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................AGGACGAAGATGACGAGGAGGAGCCT............................................................... | 26 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................C---GA---GT--------CGG---A---TGATGATGAGGA..................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................GGACGAAGATGACGAGGAGG.................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................ATGATG---------------ATGAATCC---GA---GT--------CGG---A---................................................................................................. | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....AA-GCTGA-GG---------------------------AGGATGATGA------T------GA.................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................GTG------AGGC---TAA---TGTTAC---TA---------AGCCAAT............................................................................................................................................................................................. | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................A---TGTTAC---TA---------AGCCAATCGC.......................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................................................GACGAAGATGACGAGGAGGAGCCTG.............................................................. | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................GATGATGATGATGGTG------AGGC---T............................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......A-GCTGA-GG---------------------------AGGATGATGA------T------GAG................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................TGATGGTG------AGGC---TAA---TGT..................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................AATCC---GA---GT--------CGG---A---TGATGATG......................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................................G---A---TGATGATGAGGACGAAG................................................................................ | 19 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................GATGATGAGGACGAAGATGACGA......................................................................... | 23 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................................................................................................GACGAAGATGACGAGGAG..................................................................... | 18 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............G---------------------------AGGATGATGA------T------GAGGAAGA.............................................................................................................................................................................................................................................................................. | 20 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........TGA-GG---------------------------AGGATGATGA------T------GAG................................................................................................................................................................................................................................................................................... | 19 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................TGATGATGATGGTG------AGGC---.............................................................................................................................................................................................................................. | 18 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................ATGATGATGATGGTG------AGGC---.............................................................................................................................................................................................................................. | 19 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................GAAGATGACGAGGAGGAG.................................................................. | 18 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................TCC---GA---GT--------CGG---A---TGATGGAA......................................................................................... | 19 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |