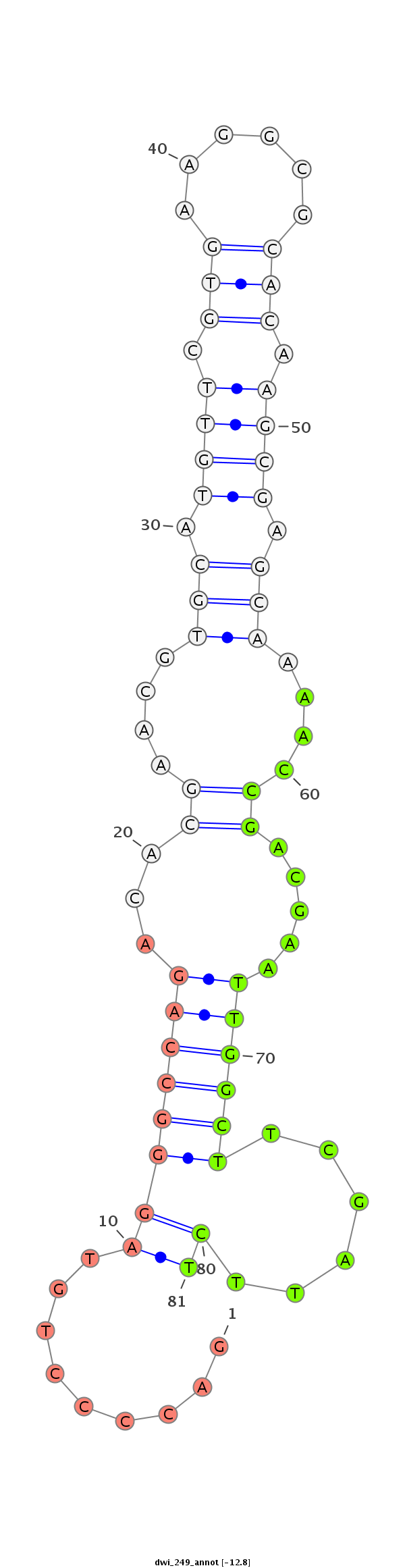
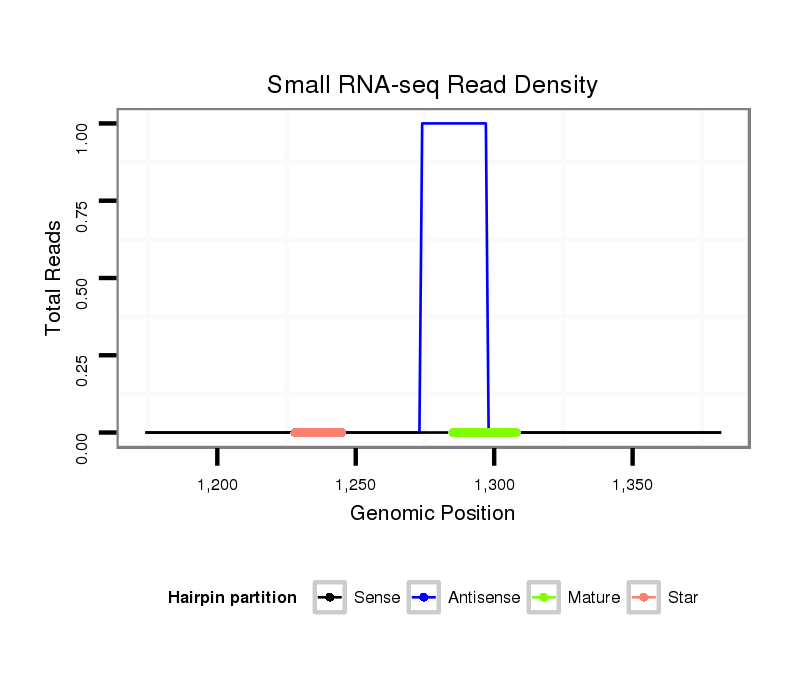
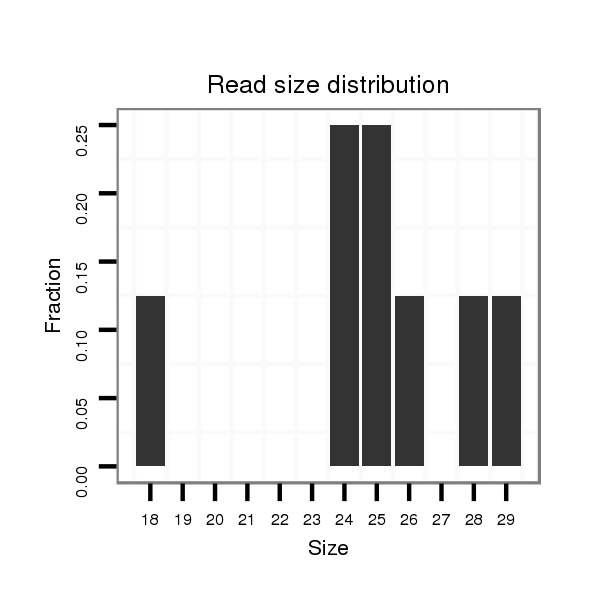
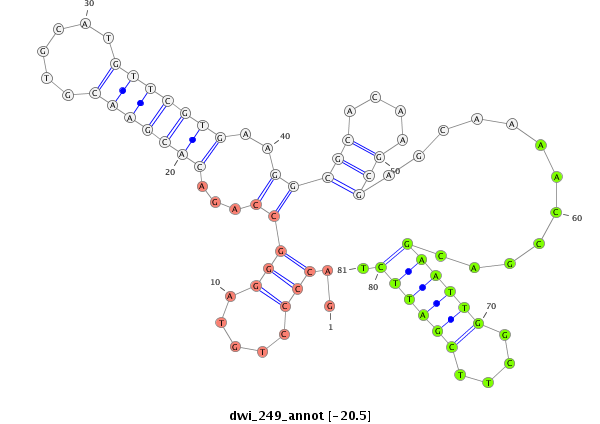
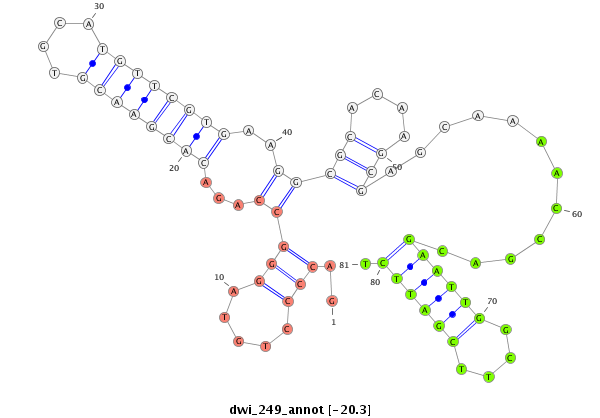
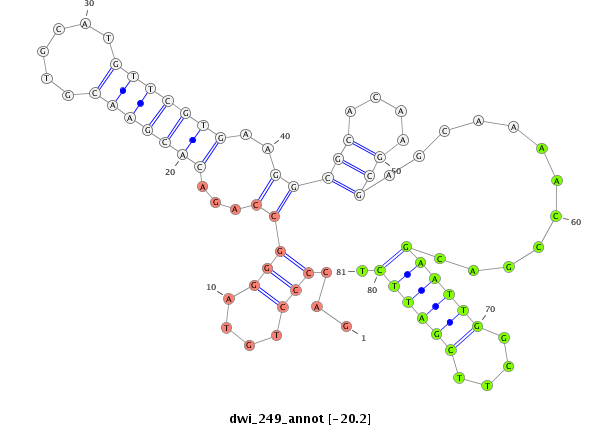
| GCTGTG-AGCG-----------------------------TGGCGGAGAGCGTCGCGGGAAGCGCCGCTG-TGAGCGTGGCAGAGAGCGTCGCGGGAAGAGAGAGTGAGCAGGAGGAGC------------------------------------------------GGGTG-------------------------TCGGACGAGTTGACCGAAATTACGAAGCA-G-----ATGACGTTGTATACAGCACGAAAGGATCTCGCGGAGTTGAAAGCGAAGGT---------------TGCCGC-CTTGGAAGGC | Size | Hit Count | Total Norm | Total | M026
Head | M043
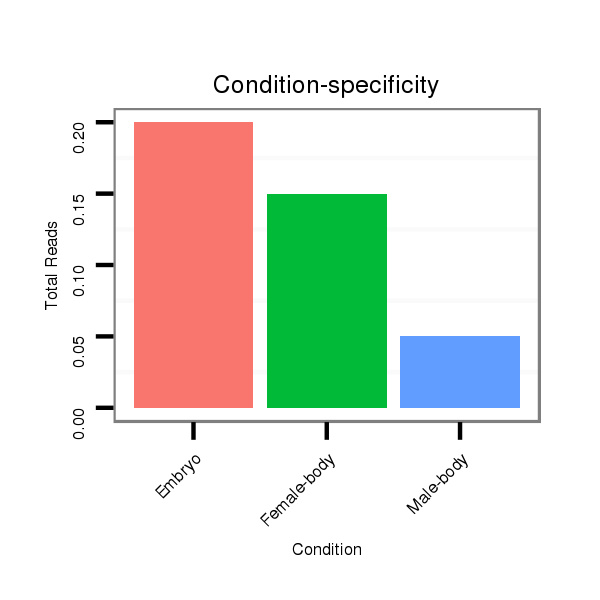
Female-body | M056
Embryo | V046
Embryo | V052
Head | V058
Head | V120
Male-body | SRR1275488
Male larvae | SRR1275486
Male prepupae | SRR1275484
Male prepupae | GSM1528802
follicle cells |
| ...................................................................................................................................................................................................................................G-----ATGACGTTGTATACAGCACGAAAGGA........................................................ | 27 | 5 | 2.00 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 |
| ..............................................................................................................................................................................................................................................................................TGAAAGCGAAGGT---------------TGCCGC-CTTGGAA... | 26 | 8 | 1.25 | 10 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 |
| .............................................................................................................................................................................................................................................CGTTGTATACAGCACGAAAGGATC...................................................... | 24 | 6 | 1.17 | 7 | 0 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................TGACGTTGTATACAGCACGAAAGGATCT..................................................... | 28 | 5 | 1.00 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| ...................................................................................................................................................................................................................................G-----ATGACGTTGTATACAGCACGAAAGG......................................................... | 26 | 5 | 0.80 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| .............................................................................TGGCAGAGAGCGTCGCGGGAAGAGA..................................................................................................................................................................................................................... | 25 | 5 | 0.40 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................TGAGCGTGGCAGAGAGCGTCGCGGG........................................................................................................................................................................................................................... | 25 | 5 | 0.40 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................TGAAAGCGAAGGT---------------TGCCGC-CTTGG..... | 24 | 8 | 0.25 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TG-TGAGCGTGGCAGAGAGCGTCGCG............................................................................................................................................................................................................................. | 25 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................GTGGCAGAGAGCGTCGCGGGA.......................................................................................................................................................................................................................... | 21 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................TCGCGGAGTTGAAAGCGAAGGT---------------TGC.............. | 25 | 10 | 0.20 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................CGAAGCA-G-----ATGACGTTGTATACAG.................................................................. | 24 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................GTGGCAGAGAGCGTCGCGGGAAGAG...................................................................................................................................................................................................................... | 25 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................GTTGTATACAGCACGAAAGGATCTCG................................................... | 26 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................GTGGCAGAGAGCGTCGCGGGAA......................................................................................................................................................................................................................... | 22 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................CGTTGTATACAGCACGAAAGGATCTCGC.................................................. | 28 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................GC------------------------------------------------GGGTG-------------------------TCGGACGAGTTGACCGT..................................................................................................... | 24 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................TGGCGGAGAGCGTCGCGGGAAGCGCC......................................................................................................................................................................................................................................................... | 26 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................GTTGACCGAAATTACGAAGCA-G-----ATGAC............................................................................. | 27 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................................................................................CGTTGTATACAGCACGAAAGGATCTC.................................................... | 26 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................GTTGTATACAGCACGAAAGGATC...................................................... | 23 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TCGCGGGAAGCGCCGCTG-TGAGC............................................................................................................................................................................................................................................... | 23 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................CGTTGTATACAGCACGAAAGGATA...................................................... | 24 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................CGTTGTATACAGCACGAAAGGA........................................................ | 22 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................CGTTGTATACAGCACGAAAGGAT....................................................... | 23 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................CGTTGTATACAGCACGAAAGGATCT..................................................... | 25 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................AGTTGACCGAAATTACGAAGCA-G-----A................................................................................. | 24 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................AGAGTGAGCAGGAGGAGC------------------------------------------------GG.................................................................................................................................................. | 20 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................GAGAGAGTGAGCAGGAGGAGC------------------------------------------------G................................................................................................................................................... | 22 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................TTGAAAGCGAAGGT---------------TGCCGC-CTTGGAA... | 27 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................TGAAAGCGAAGGT---------------TGCCGC-CTTGGAAG.. | 27 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................GTTGAAAGCGAAGGT---------------TGCCGC-CT........ | 23 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................GGAGTTGAAAGCGAAGGT---------------TGCCT............ | 23 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................TCGCGGAGTTGAAAGCGAAGGT---------------TGCC............. | 26 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................CGCGGAGTTGAAAGCGAAGGT---------------TGCC............. | 25 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................GGAGTTGAAAGCGAAGGT---------------TGCCGC-.......... | 24 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................GCGGAGTTGAAAGCGAAGGT---------------T................ | 21 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................GGAGTTGAAAGCGAAGGT---------------TGCCGC-C......... | 25 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |