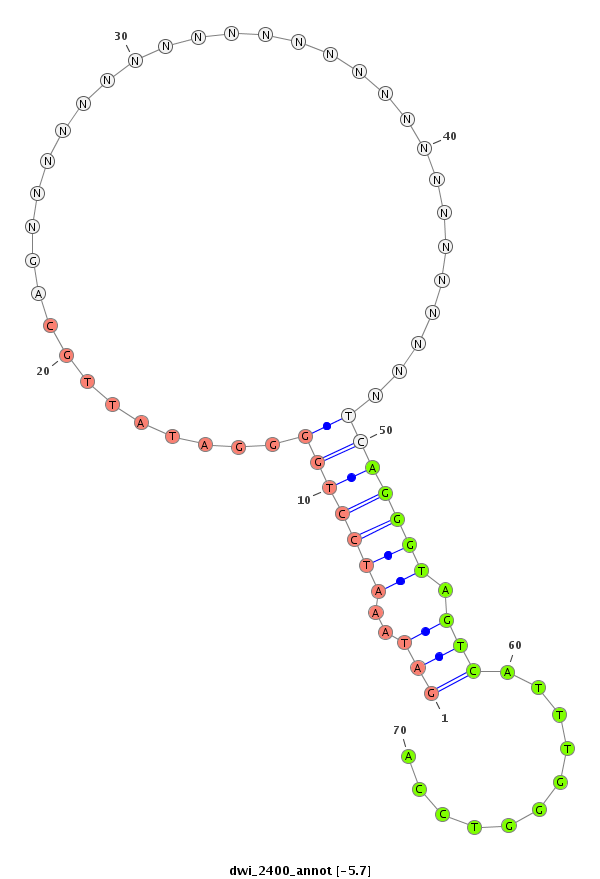
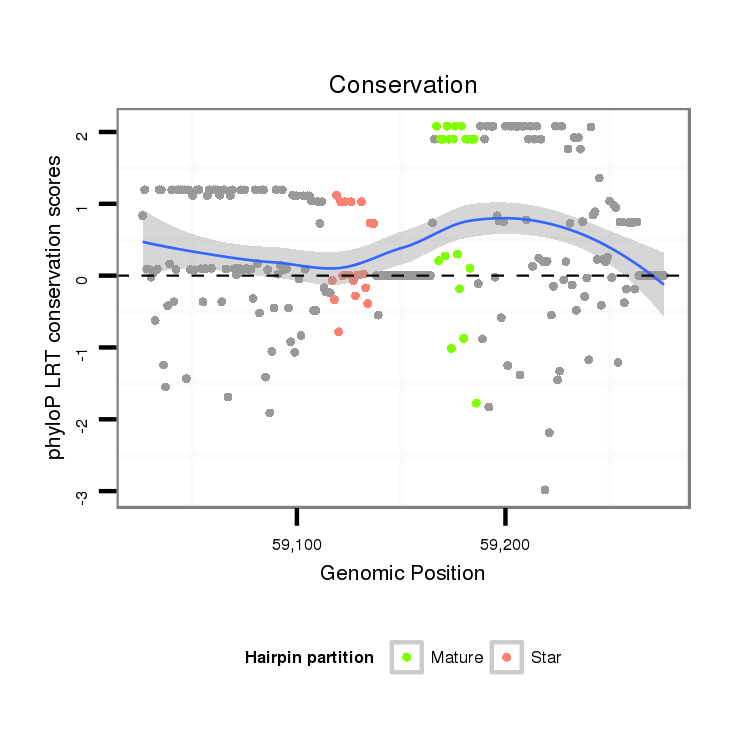
ID:dwi_2400 |
Coordinate:scf2_1100000004889:59076-59226 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -5.15 | -4.81 |
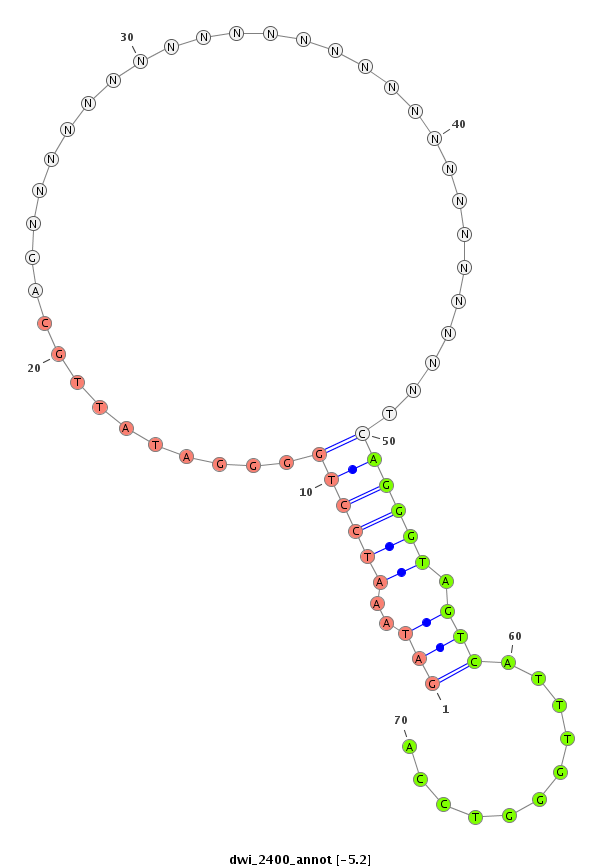 |
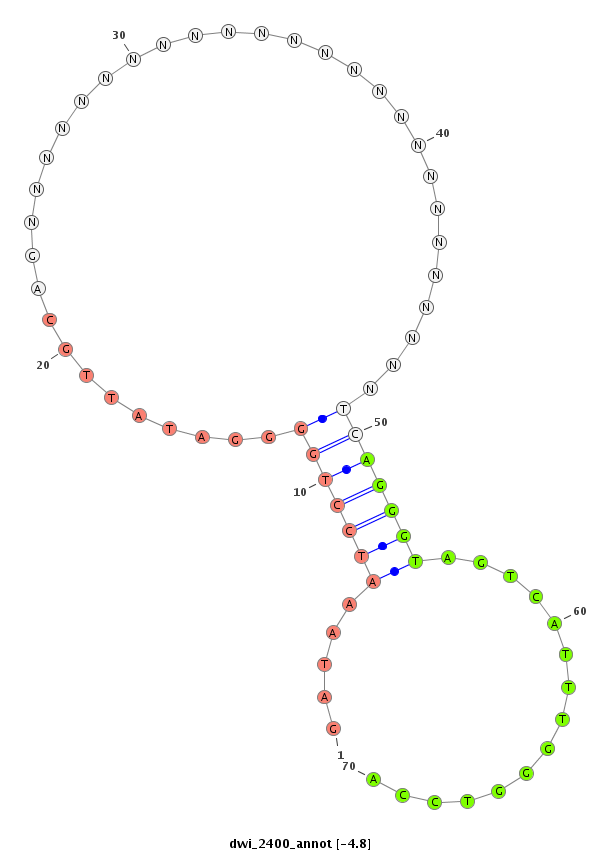 |
CDS [Dwil\GK20617-cds]; exon [dwil_GLEANR_3406:2]; intron [Dwil\GK20617-in]
| Name | Class | Family | Strand |
| CR1-1_DWil | LINE | CR1 | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTTAACGTTTAATATATATTTGTTGAATCATCTCTTAAATTAGAGCCTAACAACAGTTTGAGATATTTTCTTAGTCTAGTACTAACAATAAGATAAATCCTGGGGATATTGCAGNNNNNNNNNNNNNNNNNNNNNNNNNTCAGGGTAGTCATTTGGGTCCATTTCTATTCACTCTTTTTATAAATGATCTTCCATTGGTAGTGACACATTCTTGGGCAAACGCAAGCAGTTTGCCGCAATTCTAGGTCAAG *******************************************************************************************(((..(((((((....................................))))))).)))...........****************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V118 embryo |
V119 head |
|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................CATTCTCGGGCCAACGCGA.......................... | 19 | 3 | 9 | 0.33 | 3 | 3 | 0 | 0 |
| ...............................................................................................................................................................................................CCATTGGTAGTGACACATTCT....................................... | 21 | 0 | 20 | 0.10 | 2 | 1 | 0 | 1 |
| .............................................................................................................................................................................................................................GCAAGCACTTCGCCGCAAC........... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .............................................................................................................................................AGGGTAGTCATTTGGGTCCA.......................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ...........................................................................................GATAAATCCTGGGGATATTGC........................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ..................................................................................................................................................................................TATAAATGATCTTCCATTGGTAG.................................................. | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ............................................................................................................................................................................................CTTCCATTGGTAGTGACAC............................................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ...........................................................................................................................................TCAGGGTAGTCATTTGGGTCC........................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
|
AAATTGCAAATTATATATAAACAACTTAGTAGAGAATTTAATCTCGGATTGTTGTCAAACTCTATAAAAGAATCAGATCATGATTGTTATTCTATTTAGGACCCCTATAACGTCNNNNNNNNNNNNNNNNNNNNNNNNNAGTCCCATCAGTAAACCCAGGTAAAGATAAGTGAGAAAAATATTTACTAGAAGGTAACCATCACTGTGTAAGAACCCGTTTGCGTTCGTCAAACGGCGTTAAGATCCAGTTC
******************************************************************************************(((..(((((((....................................))))))).)))...........******************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M020 head |
M045 female body |
V117 male body |
|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................TCCCATCAGTAAACCCAGGTA......................................................................................... | 21 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 |
| ...........................................................................................................................................................................GAGAAAAATATTTACTAGAAGGTAACC..................................................... | 27 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 |
| .........................................................................................................................................................................................CTAGAAGGTAACCATCACTGTGT........................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ............................................................................ATCATGATTGATGTTCCAT............................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004889:59026-59276 + | dwi_2400 | TTTAACGTTTAATATATAT---------TTGTTGAATCATCTCTTAAATTAGAGCCTAACAACAGTTTGAGATATTTTCTTAGTCTAGT---ACTAACAATAAGATAAATCCTGGGGATA--------------------------------TTGCAGNNNNNNNNNNNNNNNNNNNNNNNNNTCAGGGTAGTCATTTGGGTCCATTTCTATTCACTCTTTTTATAAATGATCTTCCATTGGTAGTGACACATTCTTGGGCA--AACGCAAGCAGTTTGCCGCAATTCTAGGTCAAG |
| droVir3 | scaffold_12736:236023-236101 + | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAGGTAGTCATCTTGGTCCGCTTCTCTTTACTCTTTTTATCAATGACCTTCCTTCGATACTGTCACACTCTTGAGTA----------------------------------- | |
| droMoj3 | scaffold_6498:1851539-1851614 + | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGGTAGCCATTTGGGTCCATTTCTCTTTTCCCTTTTTATAAATGACCTTCTT--AGTTGTA---T------ATGTGCAAATGCA--------------------------- | |
| dp5 | Unknown_singleton_531:1348-1423 + | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGGCAGCCATCTAGGCCCCTTACTTTTCACACTCTTTATTAATGACCTGCCGTGGGTATTGACATACTCTCGAG------------------------------------- | |
| droPer2 | scaffold_120:57004-57077 + | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGGCAGTCATCTGGGCCCCTTGCTCTTCACTCTGTTTATTAATGACCTGCCTTCGGTATTAACACACTCTTG--------------------------------------- | |
| droAna3 | scaffold_13260:1610202-1610304 - | ATATACATTTATCTTTTAT---------TTGTTGGATCTTCTCTTA-TTAAGAGACTAACAACAATTTTAAAAATCTTAA---TCTAAT---ACAT--AAATAGATTATTTTTGTAAACA----------------------------------------------------------------------------------------------------------------------------------------------------------T---------------------- | |
| droBip1 | scf7180000394163:3542-3634 - | TTTATAGCTTTGCTTTTAT---------TTATTGAATCTACTCTTAATTTTGAGACTAACAATAAGTTGACGAATCTTAACATTCAAATGTAACTAA----------------------------------------------------------------------------------------------------------------------------------------------------------------CAGTA----------------------------------- | |
| droKik1 | scf7180000302000:23180-23257 + | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGGTAGCCACCTCGGCCCGCTGCTATTTACATTGTTTATCAATGACCTCCAGCTGGTTCTAACGAATTCTAGTGTA----------------------------------- | |
| droFic1 | scf7180000453607:10597-10688 - | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAGGGAGCCACCTTGGCCCGCTACTATTTACCTTATTTATTAATGACCTTCCATTAGTAATAACACACTCTCGTGTACTTATGTAAGCAG---------------------- | |
| droEle1 | scf7180000490671:730-940 - | TTTA---TTTAAATTAGATCAGCTCGGGTTTTCGAATAATCTTTTAAATTGGAT-TTCAAGTTAAGTTGGAAGAACTCAGAAG---------------------GTTATTTTTAGAAACGAAGTTTCTAATATGAATAACGTAACATCTGGAGTGC-C-------------------------TCAGGGTAGTCATTTGGGCCCTTTTCTATTTACTTTATTTATCAATGATCTTCCTCATATCATAACACATTCTCGAGTA----------------------------------- | |
| droRho1 | scf7180000761931:723-820 - | TTTAAC-TTTGGCTTTTAT---------TTATTGAATCTTCTCTTAATTTTGAGACTAACAATAAGTTTACGAAACTTAACATTAAAATTTCACTAACAGTATAATA----------------------------------------------------------------------------------------------------------------------------------------------------------A----------------------------------- | |
| droBia1 | scf7180000298126:95078-95178 + | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGGTAGTCATTTGGGTCCCTTGCTGTTTACTTTGTTTATTAACGATCTTCCCTCAATCATAACACATTCTCATGTACTAACTTGACCACTTGACCGTA------------- | |
| droTak1 | scf7180000414378:16716-16805 - | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGGTAGTCATTTGGGCCCTTTGCTGTTTACTTTGTTTATTAACGATCTTCCCTCTATTGTAACACATTCTCGTGTACTAATGTAAGC------------------------ | |
| droEug1 | scf7180000408992:7563-7641 + | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGGTAGCCATTTGGGCCCTTTGCTTTTCATTTTATTTATAAATGATCTTCCATCTGTTATATCACAATCCCGGGTA----------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/18/2015 at 04:25 AM