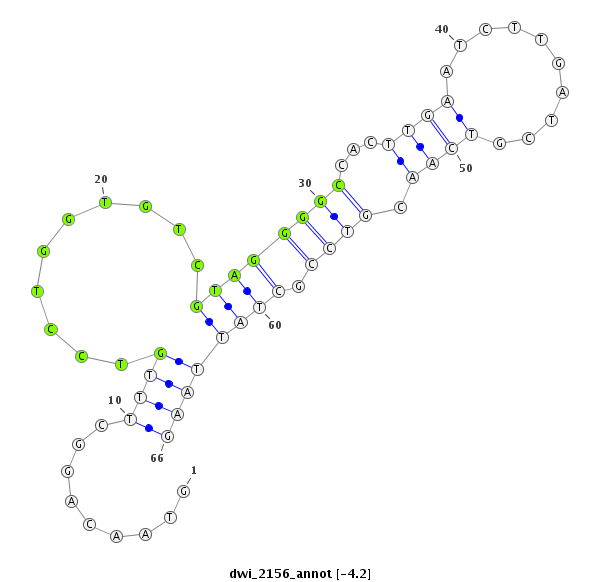

ID:dwi_2156 |
Coordinate:scf2_1100000004820:38977-39042 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -12.0 |
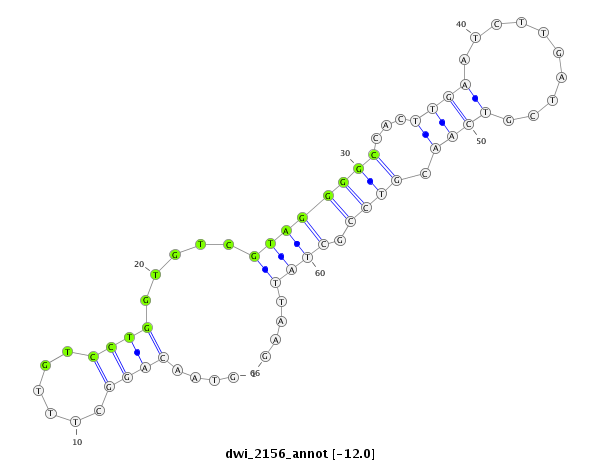 |
CDS [Dwil\GK23619-cds]; exon [dwil_GLEANR_7474:1]; exon [dwil_GLEANR_7474:2]; CDS [Dwil\GK23619-cds]; intron [Dwil\GK23619-in]
| Name | Class | Family | Strand |
| CR1-1_DWil | LINE | CR1 | + |
| ##################################################------------------------------------------------------------------################################################## AGTTCGACAGGAAGTTCATCTGGAAGAAGGGAATCTGCTTGGGCTCGCGCGTAACAGGCTTTGTCCTGGTGTCGTAGGGGCCACTTGAATCTTGATCGTCAACGTCCGCTATTAAGGGAAGAAGGGACTATAGGCAGAATTGTCGCCAGCCTGTTTTGTTGCTGAA **************************************************.........((((..........((((((((...((((..........)))).)))).))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
V118 embryo |
M020 head |
V119 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................CTCGCGCGTAACAGGCTTTGTCCT................................................................................................... | 24 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AGAAGGGACTATAGGCAGAA........................... | 20 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................GCGCGTAACAGGCTTTGTCC.................................................................................................... | 20 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 |
| ...................................TGCTTGGGCTCGCGCGTAACAGG............................................................................................................ | 23 | 0 | 20 | 0.20 | 4 | 3 | 0 | 1 | 0 | 0 |
| ..............................................................GTCCTGGTGTCGTAGGGGC..................................................................................... | 19 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| ...........................................................TTTGTCCTGGTGTCGTAGGGGCC.................................................................................... | 23 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................CTGCTTGGGCTCGCGCGTAACA.............................................................................................................. | 22 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ........................................GGGCTCGCGCGTAACAGGCTA......................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................GGGCTCGCGCGTAACAGGCT.......................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................TCCTGGTGTCGTAGGGGCC.................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| AGGTTGAGAGGAAGTTCAT................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................CTCGCGCGTAACAGGCTTTGT...................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GTAACAGGCTTCGTCCTGGTGTCGTA.......................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................TGGTGTCGTATGGGCCACTTGAAT............................................................................ | 24 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................TGCTTGGGCTCGCGCGTAACA.............................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................AATAGGGTAAGAAGGGACT..................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................AAGGGAAGAAGGGAGTAG................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................TTGGGCTCGCGCGTAACAGGCTTTG....................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................GGCTTTGTCCTGGTGTCGTAGGG....................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TTGTCCTGGTGTCGTAGGGGC..................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
|
TCAAGCTGTCCTTCAAGTAGACCTTCTTCCCTTAGACGAACCCGAGCGCGCATTGTCCGAAACAGGACCACAGCATCCCCGGTGAACTTAGAACTAGCAGTTGCAGGCGATAATTCCCTTCTTCCCTGATATCCGTCTTAACAGCGGTCGGACAAAACAACGACTT
**************************************************.........((((..........((((((((...((((..........)))).)))).))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
V118 embryo |
|---|---|---|---|---|---|---|---|---|
| ............................................................................................ACTACTAGTTGCATGCGATAATT................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .............................................................ACAGGACCACAGCATCCCCGGTGAACTT............................................................................. | 28 | 0 | 20 | 0.65 | 13 | 10 | 0 | 3 |
| ......................................................................................................................................GTCTTAACAGCGGTCGGACAAA.......... | 22 | 0 | 5 | 0.40 | 2 | 0 | 2 | 0 |
| .................................................................................................................................TATCCGTCTTTGCAGTGGTCGG............... | 22 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| .......................................................TCCGAAACAGGACCACAGCAT.......................................................................................... | 21 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 |
| ..............................................CGCGCATTGTCCGAAACAGGACCAC............................................................................................... | 25 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 |
| ....................................................................................AACTTAGAACTAGCAGCC................................................................ | 18 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................CGGTCGGGCAAACCCACGAC.. | 20 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 |
| ................................TAGACGAACCCGAGCGCGCATTGT.............................................................................................................. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .............................................GCGCGCATTGTCCGAAACAGGAC.................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .................................................................................................GAGTTGCAGGCGGTGATTC.................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004820:38927-39092 + | dwi_2156 | AGTTCGACAGGAAGTTCATCTGGAAGAAGGGAATCTGCTTGGGCTCGCGCGTAACAGGCTTTGTCCTGGTGTCGTAGGGGCCACTTGAATCTTGATCGTCAACGTCCGCTATTAAGGGAAGAAGGGACTATAGGCAGAATTGTCGCCAGCCTGTTTTGTTGCTGAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 05/18/2015 at 03:27 AM