| droWil2 |
scf2_1100000004768:2263627-2263877 + |
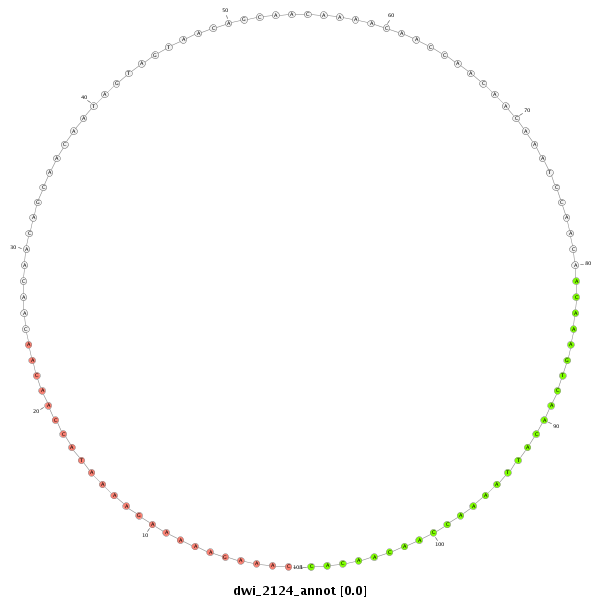
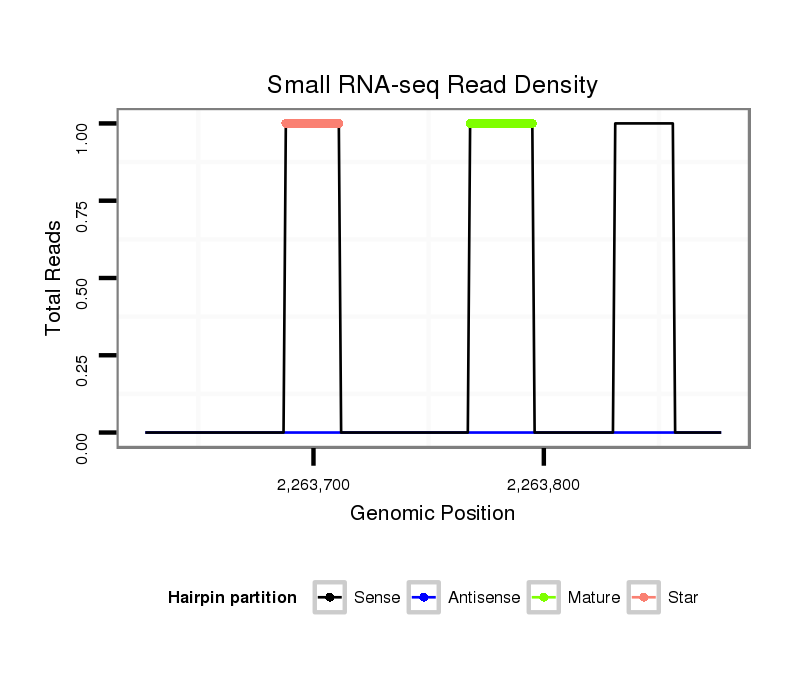
dwi_2124 |
AAAAAAACAAAAAAA----------------AAACAAAAATAAAATAA------A---ACAAAC------A-AACAAA---------------------------------------------------------------------------------------------------------CAAAAAAAAATAA------------------------------------------------------------------------------------GACAAAGA------------AA---------------------AAG-----------AAAATA------------------------CCAACA----------AC------AACAACAGCAACAATAGTA------------GT--AACAGCAACAA---------AACAAC---CAACAAC---------------------------AAATCCAACAAC-----------------------AAAGTCAACAT----TA------------------------------------------A------AACCAACAA-CAC---------TACAACAACA---------TCCACACCGGT---------------TAAACAAAC-----------------------AG---C-ACA----------------------AGAA-----------------------------------------------------------------------------------------------------------------------------------------------------------CAA---CAAC---AACTA----------C------AA---AATCCCGA-ACAA--------------GG------TGATATTCCGGAG----------------- |
| droVir3 |
scaffold_12970:7887274-7887494 - |
|
AACAAACAGACTAAC----------------AAACAAATATACAAAGC------A---GCC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCAA---------AAGTGCACAGT------------GCAATAACAACAACAACAACA-----------AC--AACAACAAC---AACAACAACAACA--------------ACA---------ACAACAACAACAACAACAA---C---------------------------------AACAACAACA-----------------------ACAACAACAA----CAACAACA---------------------------ACAACAACAAC--AACA----------------------------------------ACAACAACAAC---------------AACAACAAC-----------------------AA---CAACA----------------------ACAACAACAACAACA-------------------------------------------A----------------------------------------------------------------------------------------------------CAA---CAAC--------------------------------------------------------------AAC---AAC-----AA----------------- |
| droMoj3 |
scaffold_6473:5702156-5702408 - |
|
ACAAACACAAATACA----------------AAAGAG-------CAAAAAACAGA---------AAGAAAA-A-----------------------------AAA-AAAA--------------------------AAAAAAAACAAAACAAAGAACT---T-----------AAT----C-------------------------------------------AAATTTCC----AGCAAATCATCG-AAATGGAAT--TGAGCATCGGAATTGCCAAG---------------------------CATTTAGCGA---------GCT--------------------------A-----------------TAACAACA--GC---AGCAGC---------AACATCAACAGCAGCAGCAG--------------------------------------------------------------------------------------------------------------------------CA------------------------------ACATCA--ACAGAA----------------------------------------AC------------------------------------GGCAA--CAGCAACAGCAACAG---CAACAACAACTG-------------------------C-------------------------------------------------AACAGCAATAG----------------------------------------------------------------------------------------CAA---CCAG-------------------------A------TCAC--------------------------TAT---TC-----GAA----------------- |
| droGri2 |
scaffold_15203:1310443-1310680 - |
|
ACAACAGCAAGAACA----------------ACAGCA-------ACAACAACTGC---------A------A------------------------------------------------------------------------------------------------------------------CAACAAGAGC------------------------------------------------------------------------------------AACAGCAAGAG---CAA---CACCAACAT------CAACA---AC--------------A---------------ACA---------GCAA--CAACAACAGC--AACAACAAC---TGCAACAACAACT--------------GCA---------ACAACAACTGCAA---------CAAC---------------------------AAGAGCAACAAC-----------------------AACATCAACAA----CAACAGCA---------------------------ACAACAACAGC--AACA----------------------------------------ACAACAGCAAC---------------AACAACAGC-----------------------AA---CAACA----------------------ACAGCAACAACAACA-------------------------------------------G----------------------------------------------------------------------------------------------------CAA---CAAC--------------------------------------------------------------ACC---AAC-----AA----------------- |
| dp5 |
XR_group6:8816413-8816653 - |
|
AAAAACAACAGCACC----------------AAAAACATCGAAAATAACGCCAAC---------A------A-AAACA-----------------------------------------------------------------------------------------------------------------CCGAC------------------------------------------------------------------------------------AACAACAGAAC---CTA---GA------A------CAACA---AC--------------AACA------------------------CCAACA----------AC---AACAGC---AACATCAACAACA------------AC--A---------AACGCAACACCAAAAA---CAACAAC---------------------------AGCACCTAAAAC-----------------------AACAACAACAT----CA-----------ACA----------------------ACAACA---------------ACAAATCAAACAATAACAC------------------CAAAAAC---------------AACACAAAC-----------------------AG---CAACA----------------------CCAACAACACCAA-------------------------------------------CAA----------------------------------------------------------------------------------------------------TAT---C--------------------------------------------------------------------ACCAACAACTAAA----------------- |
| droPer2 |
scaffold_23:1625455-1625686 - |
|
CAACAAACAACAACA----------------AAACGAAGGAAG-AAAAAACCAAC---------A------AAACAAACATAGTACAAAGGATTCATCAATTTTA-CAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCA------------AA---------------------AAC-----------AAAAAC------------------------CAAGC----------------AACAGC---AACAACAACAAAT-----------------------------------------------------------ATTCATAAAGCGATAAAAAAAAATATCAAAACA-----------------------AA---------------------------AGCAAAATTTAATCAATAAA-----A---A------AATAAAAAA-AAC---------AACAACAATA---------TATATATC-------------------------------------------------------------------------TTAAAAATAC---------CA---A----------------------------------------CAA----------------------------------------------------------------------------------------------------CAACAACAAC--------------------------------------------------------------------A-------AC----------------- |
| droAna3 |
scaffold_13117:4523092-4523307 - |
|
AAAAAAAAAAAAATA----------------GATAAAAAACAAAACAA------G---AAAAAC------A-GAAAATAAAACA-------------------------------------AAAATAAAG---AAAAAAACAAACA-----GGACACGGTGAAAAA---AAGAAATG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C----------------AACAGC---AACAACATCATCA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCATC---------------AACAACAAC-----------------------AA---CATCA----------------------ACAACAGAAACAGCAACAACAACAACAACAAGTG----------TA-----------------------------------------------------------------------------------------------------------------------------------------------------G---TGTGTCGC-ACAAGTGCGACCTGTGCGGG------TGAAATAATGAAG----------------- |
| droBip1 |
scf7180000396434:620799-621047 + |
|
ACAACAACAAAAATT----------------ACAAAA-------ACAACAACAAACACAAAAA---------------------------------------TTA-CAAA-----------------------------------------------------------------------AACAACAACAAAAAC------------------------------------------------------------------------------------AACAACAA------------AA---------------------ACA-----------AAAATTACAAAAACAACAACA---------AAAA--CAACAACAAA--AACAAAAAC---C--------------------------------------A------------------CAACAACAACAAAAATTACAACA---------------------A--------------CAACAACAAAAATTACAAC--GAAACAACAAAA---------------------------A-----------------------T-TAC---------AACAACAACA---------AACACAAAAAT---------------TACAAAAAC-----------------------AA---CAACA----------------------AAAACAACAACAACA-------------------------------------------A----------------------------------------------------------------------------------------------------CAA---AAAC--------------------------------------------------------------AAC---AAC-----AA----------------- |
| droKik1 |
scf7180000302592:702839-703121 + |
|
AACAAAACAAAAAAA----------------GCCGCC-------AAAAAAATTCAAACAAAAA---------------------------------------TTG-CAAGCAAAACAAACAAAAATAAGAAACAAAAAATGAAACA-----AGAGAGAACGAAAAA--GGAGCGACGAAA----------------CAGCATCAGAAACAA-----------------------------------------------------------------------------CAG---T-----------------------------------------------------------------------------------------------------------------------------------T---------------------AAC--------------------------------------------------------------------------------------------------------------TTTGGTGTATCAA-----C---A-----ACAATTACAG-AAC---------GGCAACAAATCATCTTCCATTAGCAACGGCATTAACAATAGTAAC---------TAAAA-----C------------AA---CA----------------------------------A------------CAACGACAGCCGCTGGAGGAG--------------------------------------------------------------------------------------------------------------------GAG---GAAC---A----ACAACACAAGCAG----------CGCCCAA-CCAA--------------GGTCAACT---TT-----TGG----------------- |
| droFic1 |
scf7180000453438:40056-40320 - |
|
AACAGCAGGATCAGC----------------AGAAAGATG------ACC---AG---CAGC---A------A------------------------------CCA-CAGC-----------------------------------------------------------------------AACAACAACAAATTC------------------------------------------------------------------------------------AAGAGCAG------------CT---------------------GGG-----------AGAACT------------------------CCTAAA----------GC---AACAGC---AGCAGCAACAGCA------------GC--A---------------------AACACAGCAACGACACCAACAATTCCAACA---------------------G--------------CAACTGTGGCAACTACAACGA----CA-----------ACAGC----------------------AGAA---------------TCAGCAGAAAGATGACCAG------------------CAGCAAC---------------CACAGGAAC------AGCA--ACAATTTCAACAGCAGCAACA----------------------------------------------------ATTA----------CAAGAAAAGCTTGGA----------------------------------------------------------------------------------------------------CAA---C--------------------------------------------------------------------TGCAACAGCAGAA----------------- |
| droEle1 |
scf7180000490751:971131-971349 + |
|
AGAAACAGAAACAGA----------------ACATAAACA------TTC---GA---CAAC---A------A------------------------------CAA-CAAT-----------------------------------------------------------------------AACAATAGCAATAAC------------------------------------------------------------------------------------AAGTGCAG------------CG---------------------ACA-----------AATATA------------------------CCAACT----------GC------AGC--------------------------------------------AACAAC---------------------------------------------AACAGTAACAAC-----------------------AACAGCAACAA----CAACAGCAACAAC--AAC----------------AGCAACAACAAC--AGCA----------------------------------------ACAACAACAAC---------------AACAACAACAACAACAGCA--GCAACAACAACAG----------------------------------------------------CAACAACAAGAA----------CAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCAACGAAAAAG----------------- |
| droRho1 |
scf7180000779985:424282-424542 - |
|
AACAAAACAAATACA----------------AACAAA-------ATAAC-ATTGT---------A------A------------------------------------------------------------------------------------------------------------------------------------TAAAAAAAGC-A----------------------------------------------------------------AACCGCACGAC---AA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAGAAAAGGCAATAACAACAGAAAC---------CACCA------------------AG---CC----------------------------------A------------CAAAGCCAGCCACCGAAAGAGATAG-------------AGCGCTGAAACAA-AAAAGGCGAAATACGAAAAGTAAAAACCCTGAAAACCCCCTTTGAAAACCCCCATTGAAAACCGCCTGGAAAACCACTAGAAGCCCGC---TACAAAATGTTA----------C------AA---AACTCGAA-C--------------GCCGC------TGCCGCAGCGCAG----------------- |
| droBia1 |
scf7180000297886:2-227 - |
|
ACAAGAACAAGAACA----------------AGAACA-------AGAACAAGAAC---------A------A------------------------------------------------------------------------------------------------------------------GAACAAGAAC------------------------------------------------------------------------------------AAGAACAAGAA---CAA---GAACAAGAA---------CAAGAACA-----------------------------------------AGA-----------AC--AATAACAAT---AACAATAACAATA--------------ACA---------ATAACAATAACAATAACAA---T---------------------------------AACAATAACA-----------------------ATAACAATAA----CAATAACA---------------------------ATAACAATAAC--AATA----------------------------------------ACAATAACAAT---------------AACAATAAC-----------------------AA---TAACA----------------------ATAACAATAACAATA-------------------------------------------A----------------------------------------------------------------------------------------------------CAA---TAAC--------------------------------------------------------------AAT---AAC-----AA----------------- |
| droTak1 |
scf7180000415352:519064-519301 - |
|
ACAGCAGCAAAATCA----------------ACAGGC-------AAAGCAGCAGC---------A------A------------------------------------------------------------------------------------------------------------------AATCAGCAGA------------------------------------------------------------------------------------CAAAGCAGCAG---TTA---AATCAGACA---------CCTAAACAGCA--------------------------G-----------------G-----------CAAAACAGC---AACAGAAACAGCAGCCCAAGCAGCAG-----------------------CAAAATCAG---A---------------------------------AGCCAAAACA-----------------------GC---------------------------AAC----------------------AAA-----AT---------C-A------------------------------GC------------------------------------AGAAG--------------------------AAGCTGTCGCCAAAACAGTCGC----C-----A---G----------------------------------------TAGTAG-------------------------------------------------------------------------------------------------T-------AC---A----ACAATCAAAACCGCAACCAAGCACACCC-ACCAAA--------------GT------TAACAACCAGGAG----------------- |
| droEug1 |
scf7180000409631:95003-95243 + |
|
AAACAAACAAACAAAC---------------AAACAAACAA---------ACAAACAAACAAAC------A-AACAAA---------------------------------------------------------------------------------------------------------CA----------------------------------------------------------------------------------------------------AACAAA---CAA---ACAAACAAA---------CAAACAAA-----------CAA---------------ACAACAACAACAACA-----------AC--AACAACAAC---AACAACAACAACA--------------ACA---------ACAACAACAACAACAACAA---C---------------------------------AACAACAACA-----------------------ACAACAACAA----CAACAACA---------------------------ACAACAACAAC--AACA----------------------------------------ACAACAACAAC---------------AACAACAAC-----------------------AA---CAACA----------------------ACAACAACAACAACA-------------------------------------------A----------------------------------------------------------------------------------------------------CAA---CAAC--------------------------------------------------------------AAC---AAC-----AA----------------- |
| dm3 |
chrUextra:28738544-28738790 + |
|
ACACAAACACAAACA----------------CAAACA-------CAAAC-ACAAACACAAACA--------------------------------------------------------------------------------------------------------------------------------CAGGC------------------------------------------------------------------------------------ACAAACACAAA---CAC---AAACACAAA---------CACAAACA-----------CAA---------------AC----------------------------ACAAACACA---AACACAAACACAA--------------ACA---------CAAACACAAACACAAACAC---A---------------------------------AACACAAACACAAACACAAACACAA--------ACACAA----------------------------------------------ACACAAACACAAACACAAACAC-A-----------AACACAAACA---------CAAACACAAAC---------------GGCAAACAC-----------------------AA---ACACA----------------------AACACAAACACAAAC-------------------------------------------A----------------------------------------------------------------------------------------------------CAA---ACAC--------------------------------------------------------------AAA---CA-----CGG----------------- |
| droSim2 |
x:5371909-5372168 + |
|
CGGAAGAGACAACAA----------------CAACAACGACC--------AAGAA------------------------------------------------------------------------------------------------------------------------------------ACCAGAGCC------------------------------------------------------------------------------------AACAACCATGGTGGCACCACGA------A------CAACA---AA--------------AACA------------------------TCAGCA----------AC---CGAAGC---GGCAGCAGCAGCA--------------GCA---------GCAGCAGCAACTAGCA---CAACAACAGCAGCAACTTCCGCT---------------------G--------------CCACAACAAC------------------------------A----------------------ACAACA--ACGAGC----------------------------------------AC------------------------------------AGCAG--CAGCAGCAGCGACAG---CGACAACAACTAGCCATGCCACGCCCAC----G-----G---A----------------------------------------CAAGAG-------------------------------------------------------------------------------------------------CAA---CAGC---AA---------------------------------------------------------CAA---CA-----TGGCACATCCCATACATCCC |
| droSec2 |
scaffold_8:1950284-1950537 - |
|
AAATAATTGGCTTATTTTCAGTACAAATACAAAGCAC-------AAAAAAAAACA---------ACAAAAA-A-----------------------------TCACAAAA--------------------------AAATAAAACGAAACGAGAGCATATA---ACATAAA----------------------------------------------------------------AAC--------CCAAAA---------------------------------------------------------------------------------------------------------------------------------------------------CAAAAACGATC------GGCTCAG--------------------------------------------------------------------------------------------------------------------------CA------------------------------GAGCCA--ACG-AA----------------------------------------AA------------------------------------GCAAA--CAGCAATAGCAGCAG---GAGCAGAGGATG-------------------------C-------------------------------------------------ACCACTGATCCCGGAGAGCTCGAGACATGAGAAGCGGAA------------------------------------------------------------CA---------------------CCGAGCAGCCACCA---AGCATCGA-CTTA--------------TG-----TAGCGGTCCATGAC----------------- |
| droYak3 |
2L:17864077-17864297 - |
|
AGCAAACCAAGCAAGA---------A-----AAACAAAAACCAAATCC------A---AAAAAC------A-GAAATAGAAACA-------------------------------------GAAACAGAA---ACCTAA---AACT-----TTAGCAAACA---ACAAAAAACACCACAA----------------CAACCACATCAATAA-----------------------------------------------------------------------------CAG---A-----------------------------------------------------------------------------------------------------------------------------------G-------------------------------C---------------------------------ATTTTGTACA-----------------------ACAAGCACAG----CGAAAGCT---------------------------AGCAAAGTCCC--ACCA----------------------------------------TTATCATCATC---------------AACAACAAT-----------------------AA---TAACA----------------------ACAACAACAACAACA-------------------------------------------G----------------------------------------------------------------------------------------------------CAG---CAAC--------------------------------------------------------------AAA---AAC-----TC----------------- |
| droEre2 |
scaffold_4770:7679360-7679570 - |
|
ACAACAACAACAACA----------------ACACAA---------------------------------------------------------------------------------------------------------------------------------------------------------------CATCATCATCAACAACAACAAGCAAACAGAC-----AACAAG--------CGAAAGCAACACCGAGTGCGTTGGC-TCACCAAG---------------------------CAAC-CACAC---------ACA--------------------------A-----------------TCACACCA--AC---AACAGC---------AACAACAACA----------------------------------------------------------------------------------------------------------------GCTGCAAC--ACAAC--------------AAC----------------------ACG-----AG---------C-A------------------------------GC------------------------------------AACAA--------------------------CAGCAATTGCCACCACAGTCAC----C-----A---A----------------------------------------CAGCAG-------------------------------------------------------------------------------------------------CAG---CAAC---A--------------------------------------------------------------ACAACACATGAG----------------- |