ID:dwi_186 |
Coordinate:scf2_1100000004902:7275739-7275798 + |
Confidence:candidate |
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -32.2 | -32.1 | -32.1 |
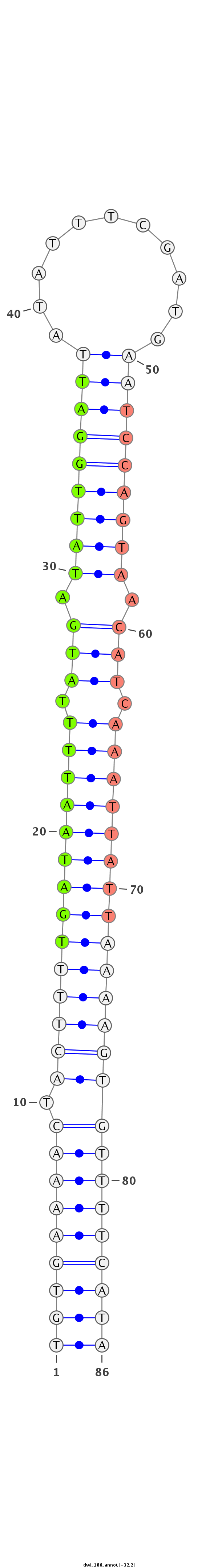 |
 |
 |
intergenic
No Repeatable elements found
|
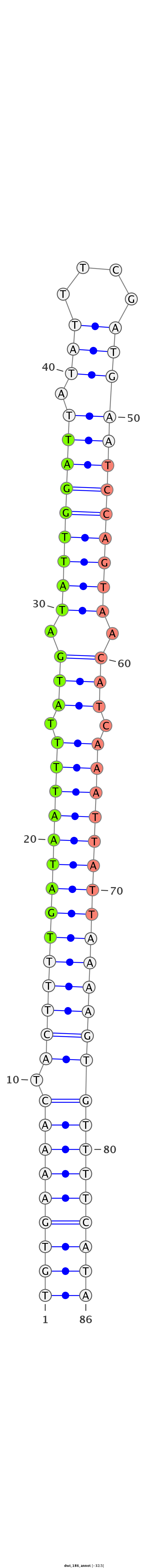
AGGCATCAATTATATTAGAAAAATATTGAGAATGATGTGAAAACTACTTTTGATAATTTTATGATATTGGATTATATTTCGATGAATCCAGTAACATCAAATTATTAAAAGTGTTTTCATAACCGAAACAAATGCAAAATTTACTCATATATGCTAAAGT
***********************************(((((((((.((((((((((((((.(((.(((((((((.(((....)))))))))))).))).)))))))))))))))))))))))*************************************** |
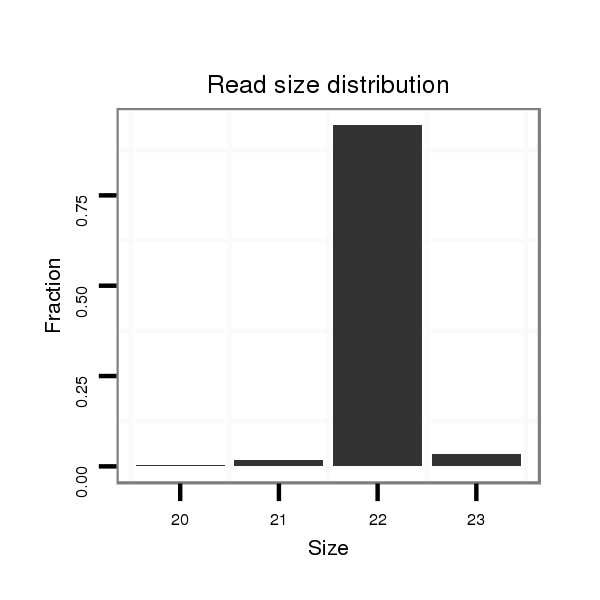
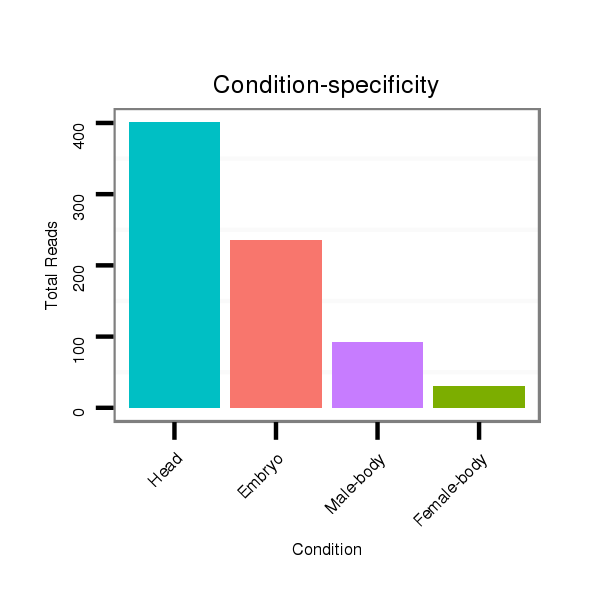
Read size | # Mismatch | Hit Count | Total Norm | Total | M020 head |
V118 embryo |
V117 male body |
M045 female body |
V119 head |
|---|---|---|---|---|---|---|---|---|---|---|
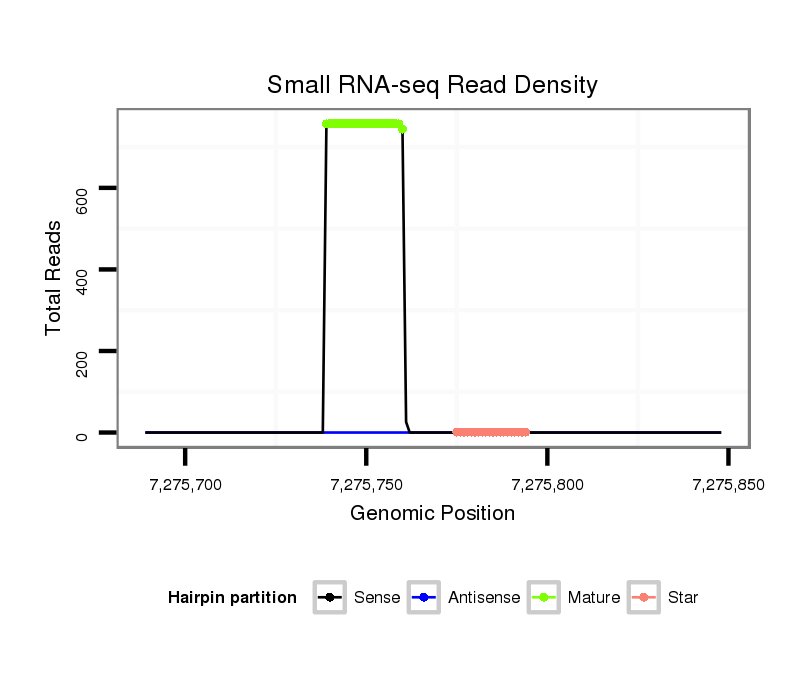
| ..................................................TGATAATTTTATGATATTGGAT........................................................................................ | 22 | 0 | 1 | 717.00 | 717 | 376 | 216 | 89 | 23 | 13 |
| ..................................................TGATAATTTTATGATATTGGATC....................................................................................... | 23 | 1 | 1 | 50.00 | 50 | 42 | 2 | 1 | 3 | 2 |
| ..................................................TGATAATTTTATGATATTGGATT....................................................................................... | 23 | 0 | 1 | 26.00 | 26 | 10 | 12 | 0 | 2 | 2 |
| ..................................................TGATAATTTTATGATATTGGATA....................................................................................... | 23 | 1 | 1 | 13.00 | 13 | 5 | 1 | 4 | 1 | 2 |
| ..................................................TGATAATTTTATGATATTGGA......................................................................................... | 21 | 0 | 1 | 13.00 | 13 | 0 | 7 | 3 | 3 | 0 |
| ..................................................TGATAATTTTATGATATTGGAC........................................................................................ | 22 | 1 | 1 | 9.00 | 9 | 9 | 0 | 0 | 0 | 0 |
| ..................................................TGATAATTTTATGATATTGGAA........................................................................................ | 22 | 1 | 1 | 4.00 | 4 | 0 | 1 | 2 | 0 | 1 |
| ..................................................TGATAATTTTATGATATTGGATCC...................................................................................... | 24 | 2 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 |
| ..................................................TGATAATTTTATGATATTTGAT........................................................................................ | 22 | 1 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..................................................TCATAATTTTATGATATTGGAT........................................................................................ | 22 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...................................................TATAATTTTATGATATTGGAT........................................................................................ | 21 | 1 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..................................................TGATAATTTTAGGATATTGGAT........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................TGATAATTTCATGATATTGGAT........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ....................AAATATGGAGTATGATGTGA........................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGATAATTTTATGAAATTGGAT........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGATAATTTTATGATATTGGCT........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGATAGTTTTATGATATTGGAT........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAATAATTTTATGATATTGGAT........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGATAATTTTATGATATTGG.......................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGATAATTTTATGATGTTGGAT........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGATAATTTTATGATATTGGATG....................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................TCCAGTAACATCAAATTATT...................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................TGATAATTTTATCATATTGGAT........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGATACTTTTATGATATTGGAT........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGATCATTTTATGATATTGGAT........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................GATAATTTTATGATATTGGAT........................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................GATAATTTTATGATATTGGATC....................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................AAATATGGCGTATGATGTGA........................................................................................................................ | 20 | 3 | 14 | 0.86 | 12 | 0 | 11 | 0 | 1 | 0 |
| ..................................................TGATAATTTTATGACATTGGAT........................................................................................ | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGATAATTTTATGACATTGGA......................................................................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................TGATAATTTTATTATATTGGAT........................................................................................ | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................TGAATTTTATGATATTGGAT........................................................................................ | 20 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................ATATTTCGGTGAGGCCAGT.................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
|
TCCGTAGTTAATATAATCTTTTTATAACTCTTACTACACTTTTGATGAAAACTATTAAAATACTATAACCTAATATAAAGCTACTTAGGTCATTGTAGTTTAATAATTTTCACAAAAGTATTGGCTTTGTTTACGTTTTAAATGAGTATATACGATTTCA
***************************************(((((((((.((((((((((((((.(((.(((((((((.(((....)))))))))))).))).)))))))))))))))))))))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
|---|---|---|---|---|---|---|
| ..........................................................................AAAAAGATACTTGGGTCAT................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droWil2 | scf2_1100000004902:7275689-7275848 + | dwi_186 | candidate | AGGCATCAATTATATTAGAAAAATATTGAGAATGATGTGAAAACTACTTTTGATAATTTTATGATATTGGATTATATTTCGATGAATCCAGTAACATCAAATTATTAAAAGTGTTTTCATAACCGAAACAAATGCAAAATTTACTCATATATGCTAAAGT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 10/20/2015 at 06:56 PM