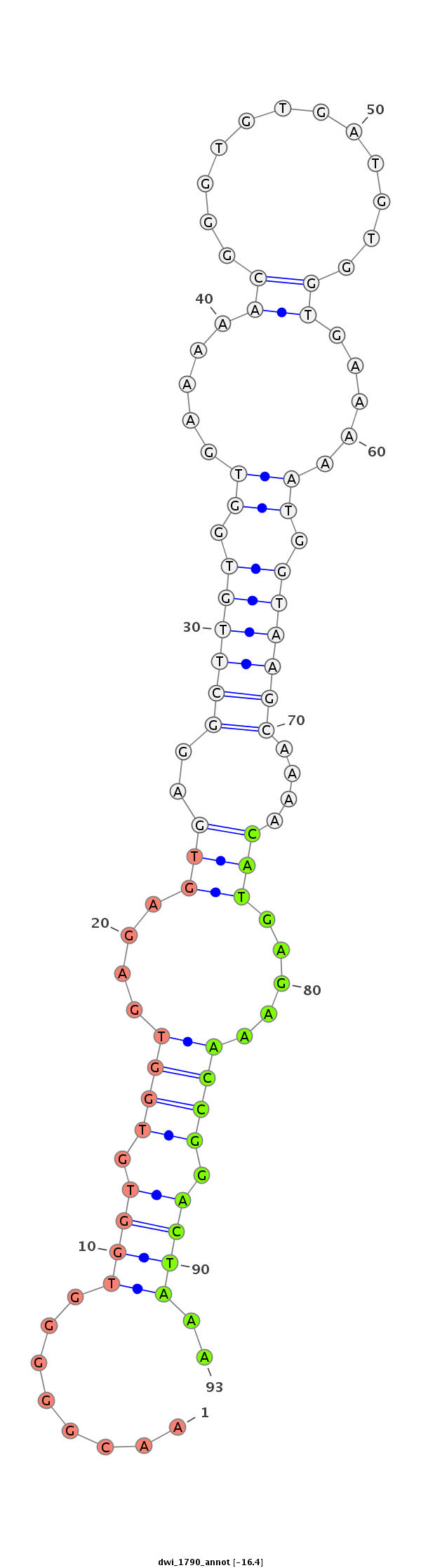
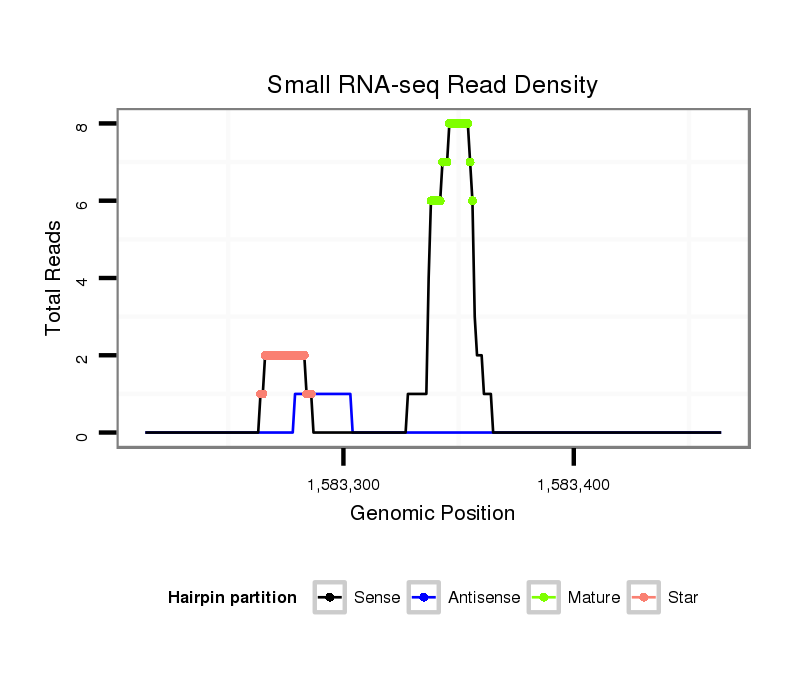
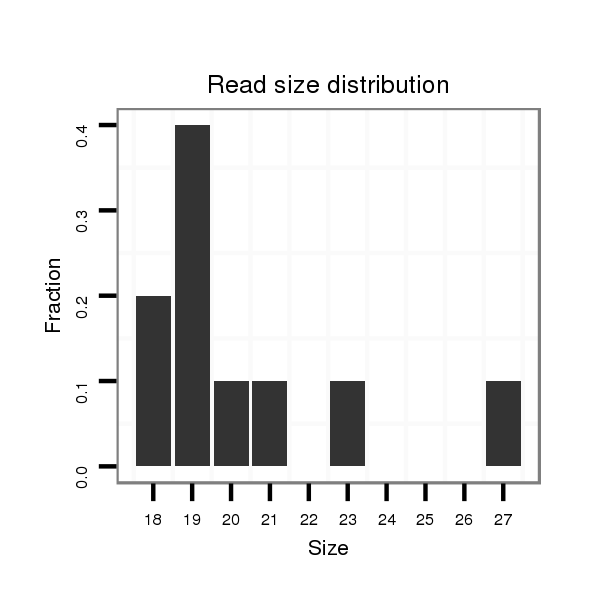
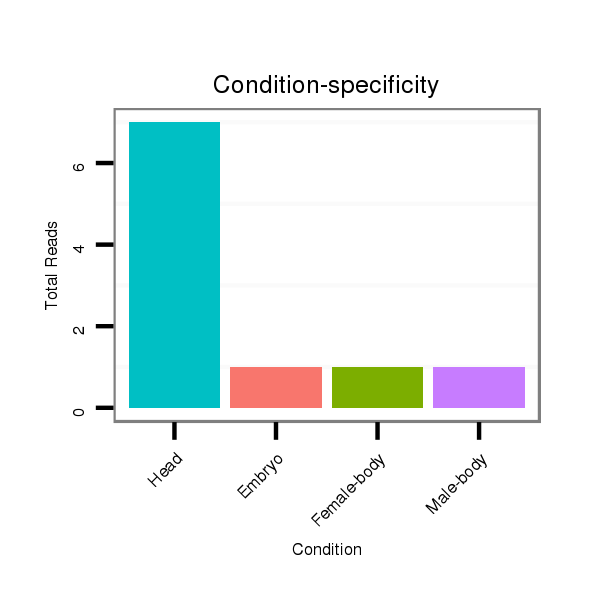
| ............................................................................................................................CATGAGAAACCGGACTAAA............................................................................................................ |
19 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
| ...........................................................................................................................ACATGAGAAACCGGACTAAA............................................................................................................ |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ..................................................AACGGGGGTGGTGTGGTGAGAGT.................................................................................................................................................................................. |
23 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
| ...........................................................................................................................ACATGAGAAACCGGACTAA............................................................................................................. |
19 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ....................................................CGGGGGTGGTGTGGTGAG..................................................................................................................................................................................... |
18 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
| ..................................................................................................................GTAAGCAAAACATGAGAAACCGGACTA.............................................................................................................. |
27 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
| .................................................................................................................................GAAACCGGACTAAAGGCA........................................................................................................ |
18 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ...........................................................................................................AAGAATGGTAAGCAAAGCATGAG......................................................................................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
| ....................................................................................................................................ACCGGACTAAAGGCAAAGT.................................................................................................... |
19 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ...........................................................................................................................ACATGAGAAACCGGACTAAAG........................................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ......................................................................................................................GCAAAACATGAGAAACCGGACTAC............................................................................................................. |
24 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ....................................................................................TGAGAAAAGGGTGTGATGCGG.................................................................................................................................................. |
21 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
| ...........................................................................GGCCTGTGTTGAAAAACGCG............................................................................................................................................................ |
20 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
| .........................................................GTGGGGTGGTTAGAGTGAGGG............................................................................................................................................................................. |
21 |
3 |
3 |
0.33 |
1 |
0 |
1 |
0 |
0 |
| ...........................................................CGTGTGGTGTGAGTGAGG.............................................................................................................................................................................. |
18 |
2 |
9 |
0.11 |
1 |
0 |
1 |
0 |
0 |
| .................................................TAACGGTAGTGGCGTGGTG....................................................................................................................................................................................... |
19 |
3 |
11 |
0.09 |
1 |
0 |
0 |
0 |
1 |
| ............................................................................GTTTTTGGTGAAAACCGGGT........................................................................................................................................................... |
20 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |