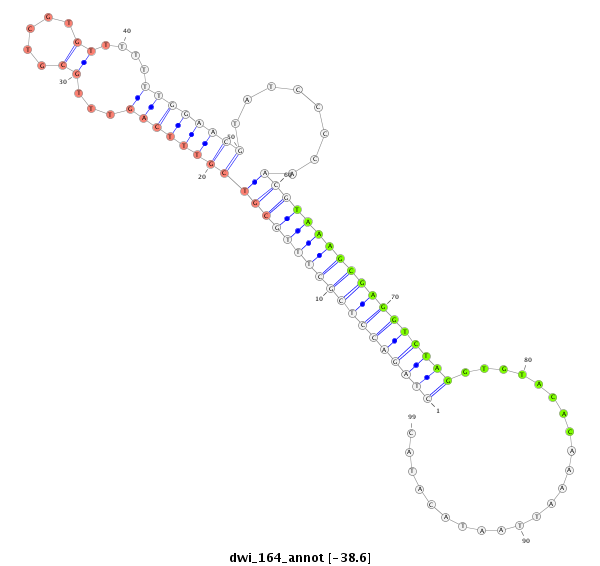
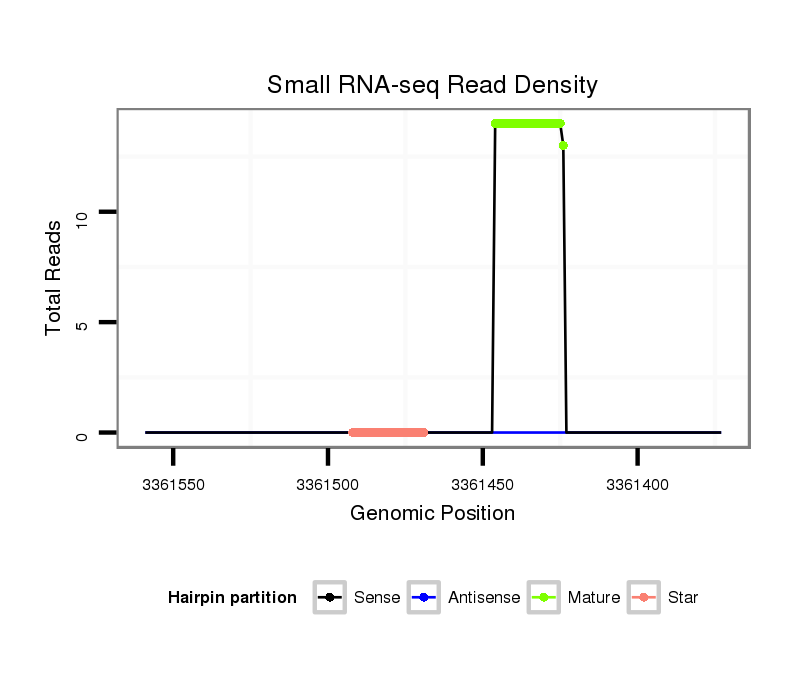
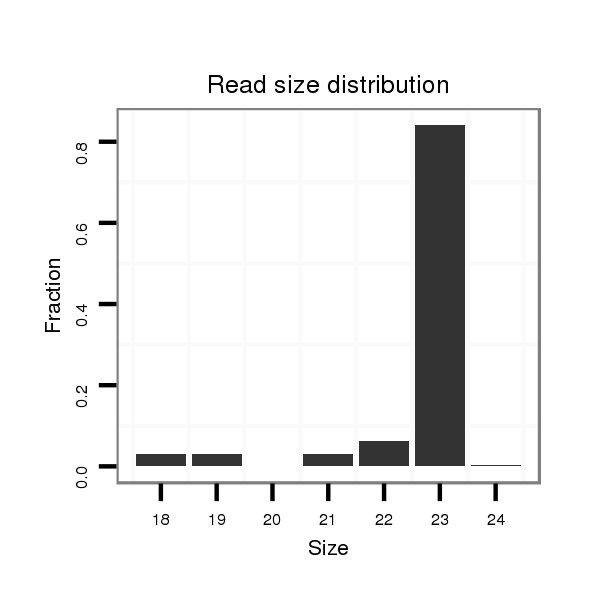
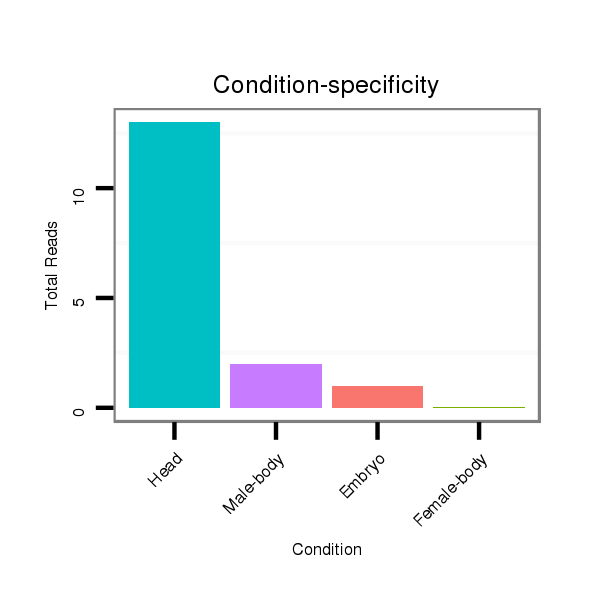
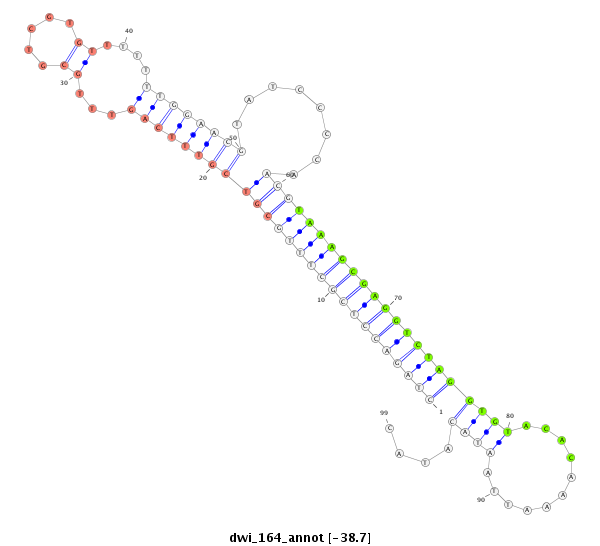
ID:dwi_164 |
Coordinate:scf2_1100000004514:3361423-3361509 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
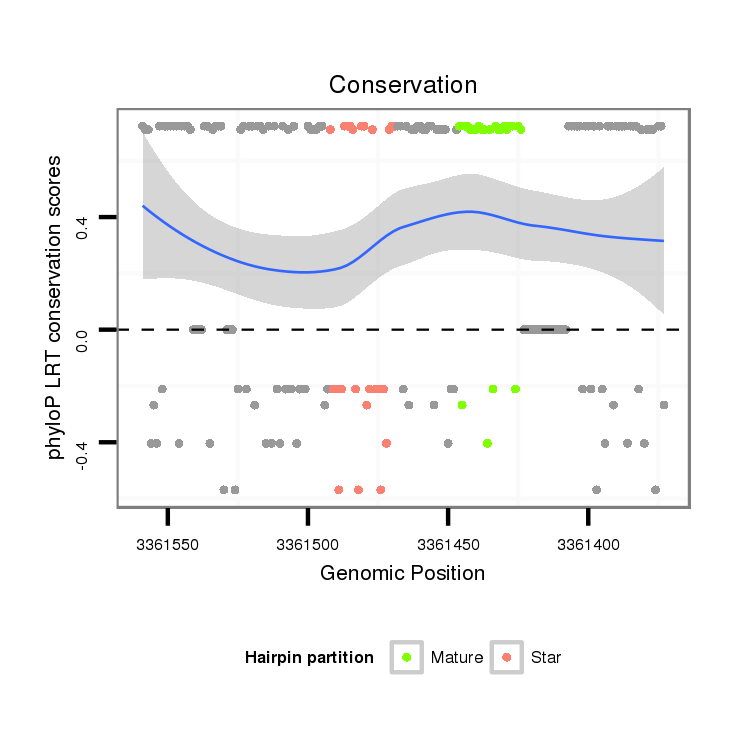
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -39.2 | -38.9 | -38.7 |
 |
 |
 |
intergenic
No Repeatable elements found
|
TGCATCTCTTATTGTTAGTTATAAAGTAAGTTTGTCTCATATTGGAATACACCTAGACCTCGCTTTGCGTCGTTTCAGTTTGCGTCGTGTTTTTTTGGAACGTATCCCCAACGTAAAGCGAGGTCTAGGTGTACACAAAATTAATACATACTATTTACATGAGTAGAATAAATCTTTGGACCCGAAT
****************************************************((((((((((((((((((((((((((...((.....))....))))))))........)))))))))))))))))).......................************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V119 head |
M045 female body |
V117 male body |
V118 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................TAAAGCGAGGTCTAGGTGTACAC................................................... | 23 | 0 | 1 | 13.00 | 13 | 11 | 0 | 2 | 0 |
| .GCAACCCTTATTGTTAGTT....................................................................................................................................................................... | 19 | 2 | 3 | 3.33 | 10 | 0 | 10 | 0 | 0 |
| .................................................................................................................TAAAGCGAGGTCTAGGTGTACA.................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| CGCAACCCTTATTGTTAGTT....................................................................................................................................................................... | 20 | 3 | 13 | 0.54 | 7 | 0 | 5 | 2 | 0 |
| ................................................................................................................GTAAAGCGAGGTCTAGGTGTA...................................................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................TAAAGCGAGGTCTAGGTGTAAAC................................................... | 23 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................AACGTAAAGCGAGGTCTAGGTGT....................................................... | 23 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................TAAAGCGAGGTCTAGGTGT....................................................... | 19 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................TAAAGCGAGGTCTAGGTG........................................................ | 18 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .GCAACCCTTATTGTTAGT........................................................................................................................................................................ | 18 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 |
| ...............................................TACACCTAGACCTCGCTTTGCG...................................................................................................................... | 22 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| ..............................................ATACACCTAGCCCTCGCCTTCCGT..................................................................................................................... | 24 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ...................................................................CGTCGTTTCAGTTTGCGTCGTGTT................................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................TTTGCGTCGTGATTTTTTGGAACG..................................................................................... | 24 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
ACGTAGAGAATAACAATCAATATTTCATTCAAACAGAGTATAACCTTATGTGGATCTGGAGCGAAACGCAGCAAAGTCAAACGCAGCACAAAAAAACCTTGCATAGGGGTTGCATTTCGCTCCAGATCCACATGTGTTTTAATTATGTATGATAAATGTACTCATCTTATTTAGAAACCTGGGCTTA
************************************((((((((((((((((((((((((((...((.....))....))))))))........)))))))))))))))))).......................**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V119 head |
V118 embryo |
M020 head |
V117 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................TTGCATTTCGCTCCAGATCCA......................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................CAAACGCAGCACAAAAAAACCTTGCAT................................................................................... | 27 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ....................................................................................AGCACAAAAAAACCTTGCATAGGGGT............................................................................. | 26 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| ...................................................................................................................TTCGCTCCAGATCCACATGT.................................................... | 20 | 0 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................GCAAAGTCAAACGCAGCACAAAAAAA........................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................AGCACAAAAAAACCTTGCATA.................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................GCAGCAAAGTCAAACGCAGCACAAAAA............................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGGATCTGGAGCGAAACGCAG.................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................GAAACGCAGCAAAGTCAAACGCAGCAC.................................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................AGCAAAGTCAAACGCAGCACA................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................AGCAAAGTCAAACGCAGCACAAAAA............................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................GTGGATCTGGAGCGAAACGCAGC................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................CAGCAAAGTCAAACGCAGCACAAAAC............................................................................................. | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................ACGCAGCACAAAAAAACCTTGCAT................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................CCACATGCGAGTTAATTAT......................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................AAAGTCAAACGCAGCACAAAAAAACCTT....................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................TATGTGGATCTGGAGCGAAACGCAG.................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................CGCAGCAAAGTCAAACGCAGCACAAAA.............................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004514:3361373-3361559 - | dwi_164 | TGCATCTCTT--ATTGTTAGTTATAAAGTAAGTTTGTCTCATATTGGAATACACCTAGACCTCGCTTTGCGTCGTTTCAGTTTGCGTCGTGTTTTTTTGGAACGTATCCCCAACGTAAAGCGAGGTCTAGGTGTACACAAAATTAATACATACTATTTACATGAGTAGAATAAATCTT-----------TGGACCCGAAT |
| droKik1 | scf7180000302476:2334781-2334957 - | TGCCAATTTTGAATTTTTAG----AACGTAAC---CCCTTATTTTGTACTGAATCCATGTTTCGCTTAACACGATTTCGCTTAACACGAGGTTTTCTAGGAACGTAACCCCCGTGTTAAGCGAGGGCCAGGTGTATAC----------------ATTTATATAACTGTAAAAAATATTAGTTTATAAATTAGCCCCCAAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 05/15/2015 at 02:53 PM