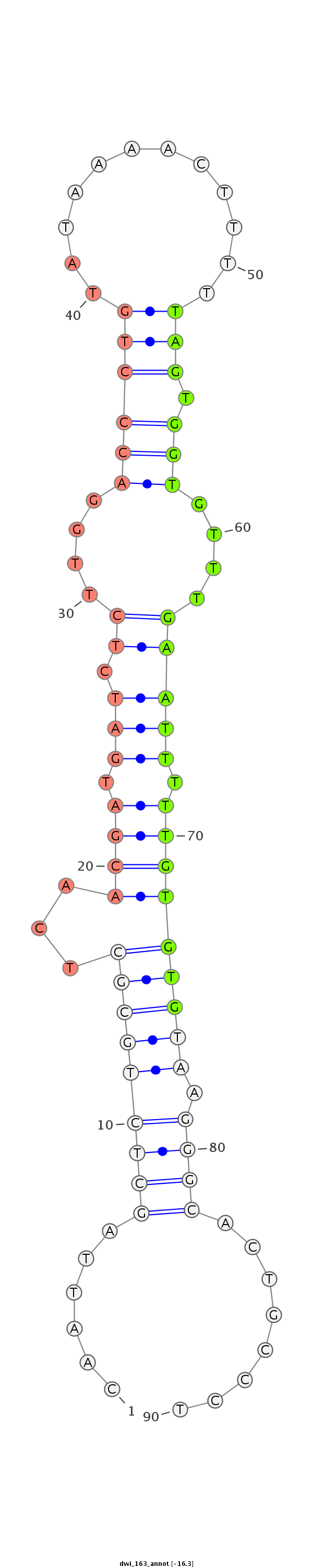
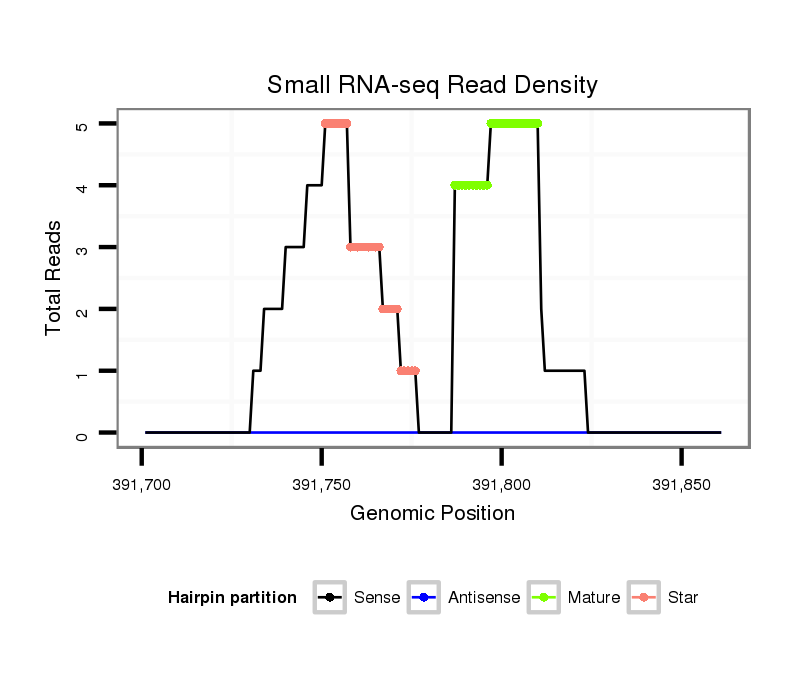
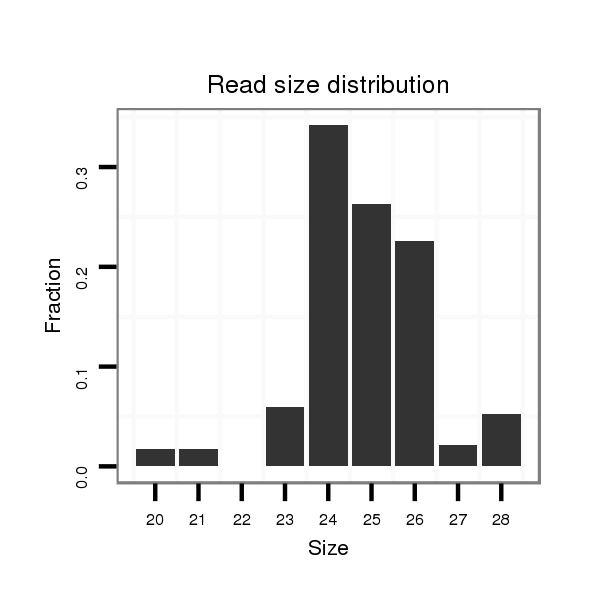
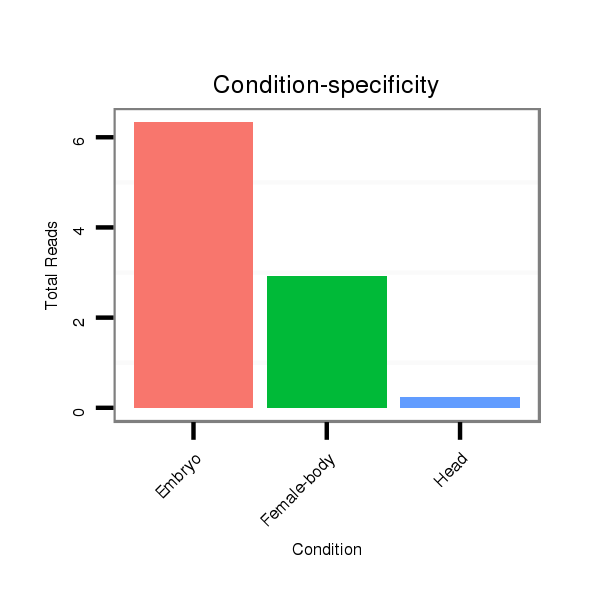
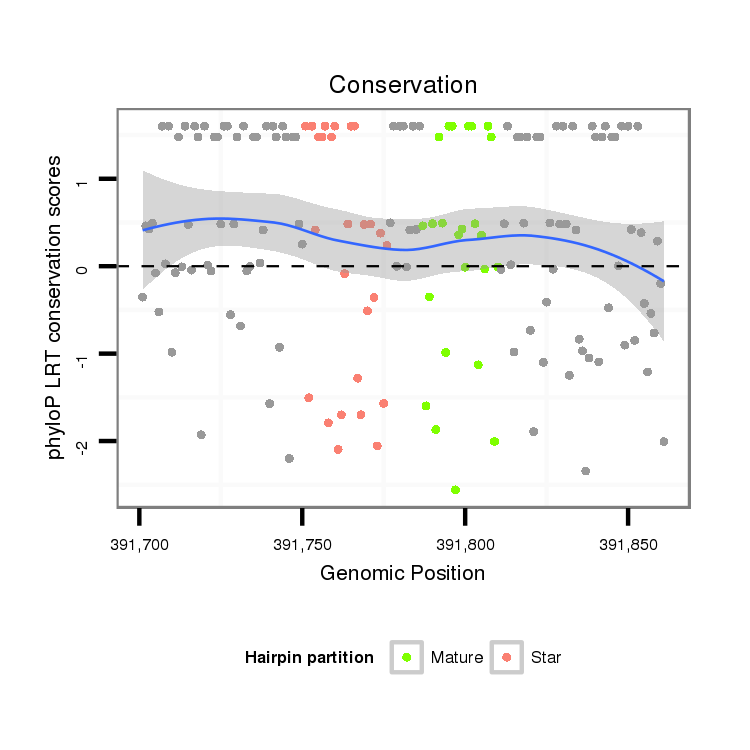
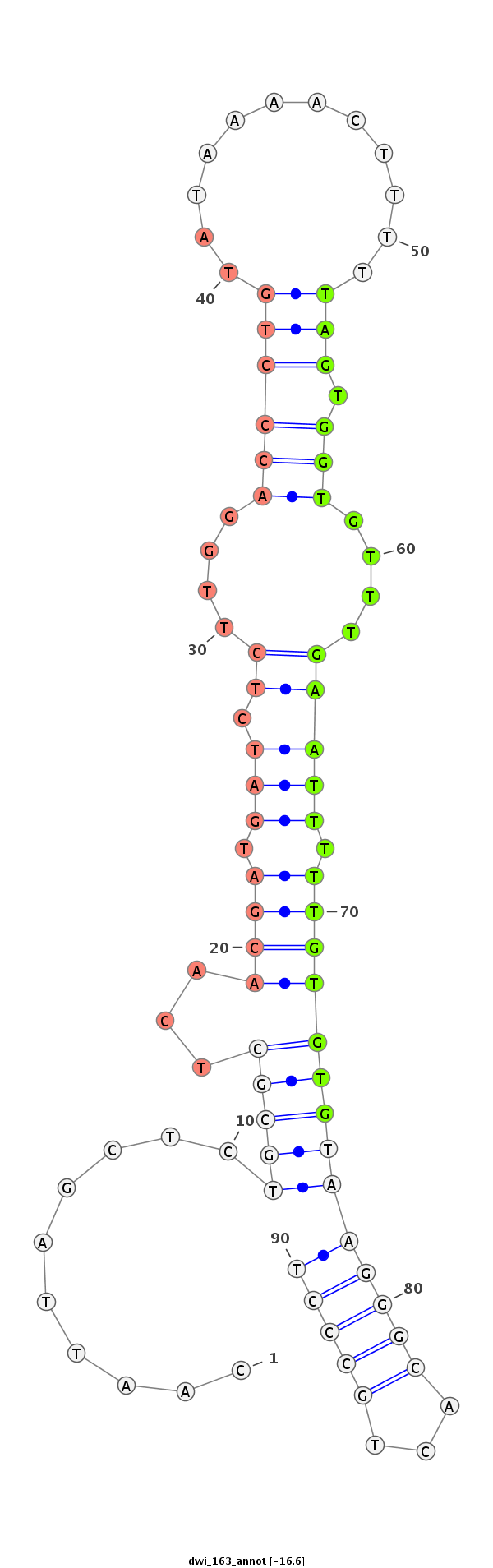
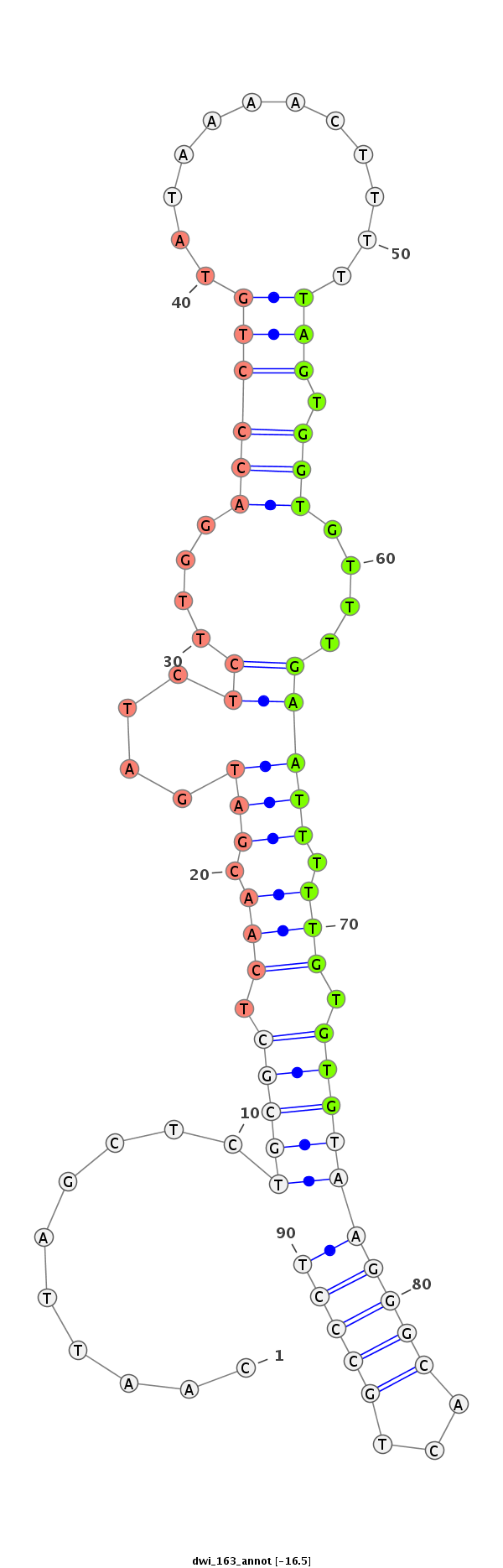
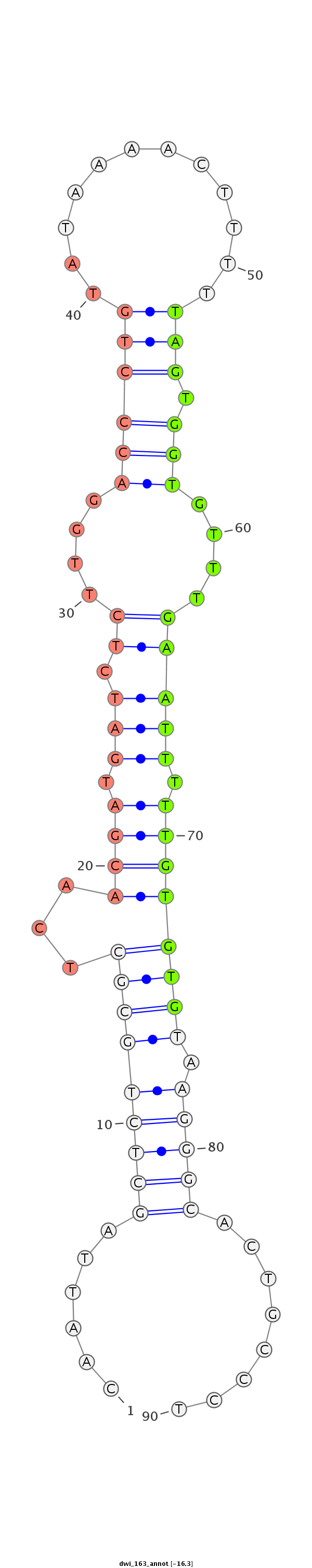
| ......................................................................................TAGTGGTGTTTGAATTTTTGTGTG................................................... |
24 |
0 |
5 |
3.00 |
15 |
12 |
3 |
0 |
| ......................................................................................TAGTGGTGTTTGAATTTTTGTGTGT.................................................. |
25 |
0 |
5 |
1.80 |
9 |
7 |
2 |
0 |
| ..................................................TCAACGATGATCTCTTGGACCCTGTA..................................................................................... |
26 |
0 |
2 |
1.50 |
3 |
2 |
1 |
0 |
| ........................................................................................................TCTGGGTAAGGTCACTGCCC..................................... |
20 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
| .................................AGCAATTAGCTCTGCGCTCAACGA........................................................................................................ |
24 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
| .......................................TAGCTCTGCGCTCAACGATGATCTCTT............................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
| ..............................TATAGCAATTAGCTCTGCGCTCAACGA........................................................................................................ |
27 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
| ................................................................................................TGAATTTTTGTGTGTAAGGGCACTGCC...................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
| .............................................TGCGCTCAACGATGATCTCTTGGACC.......................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
| ......................................................................................TAGTGGTGTTTGAATTTTTGTGTGTA................................................. |
26 |
0 |
5 |
0.60 |
3 |
2 |
1 |
0 |
| ...................................................CAACGATGATCTCTTGGACCCTGTA..................................................................................... |
25 |
0 |
4 |
0.50 |
2 |
0 |
2 |
0 |
| ......................................................................................TAGTGGTGTTTGAATTTTTGTGT.................................................... |
23 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
| .....................................................................................TTAGTGGTGTTTGAATTTTTGTGTGT.................................................. |
26 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
| ......................................................................................TAGTGGTGTTTGAATTTTTGTGTGTAA................................................ |
27 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
| ......................................................................................TAGTGGTGTTTGAATTTTTGTGTGTTT................................................ |
27 |
2 |
5 |
0.40 |
2 |
1 |
1 |
0 |
| .........................................................................................TGGTGTTTGAATTTTTGTGTGTA................................................. |
23 |
0 |
6 |
0.33 |
2 |
2 |
0 |
0 |
| ...................................................CAACGATGATCTCTTGGACCCTGTATAA.................................................................................. |
28 |
0 |
4 |
0.25 |
1 |
0 |
1 |
0 |
| .....................................................ACGATGATCTCTTGGACCCTGTATAAAA................................................................................ |
28 |
0 |
4 |
0.25 |
1 |
0 |
1 |
0 |
| ....................................................AACGATGATATCTTGGACCCTGTATAA.................................................................................. |
27 |
1 |
4 |
0.25 |
1 |
0 |
1 |
0 |
| ......................................................CGATGATCTCTTGGACCCTGTATAAA................................................................................. |
26 |
0 |
4 |
0.25 |
1 |
0 |
1 |
0 |
| ....................................................AACGATGATCTCTTGGACCCTGTA..................................................................................... |
24 |
0 |
4 |
0.25 |
1 |
0 |
0 |
1 |
| .......................................................................................AGTGGTGTTTGAATTTTTGTGTGTA................................................. |
25 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
| .....................................................................................ATAGTGGTGTTTGAATTTTTGTGTGT.................................................. |
26 |
1 |
5 |
0.20 |
1 |
0 |
1 |
0 |
| .....................................................................................TTAGTGGTGTTTGAATTTTTGTGTG................................................... |
25 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
| ...................................................................................TTTTAGTGGTGTTTGAATTTTTGTGTG................................................... |
27 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
| .....................................................................................TTAGTGGTGTTTGAATTTTTGTG..................................................... |
23 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
| .........................................................................................TGGTGTTTGAATTTTTGTGTG................................................... |
21 |
0 |
6 |
0.17 |
1 |
0 |
1 |
0 |
| .........................................................................................TGGTGTTTGAATTTTTGTGT.................................................... |
20 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |