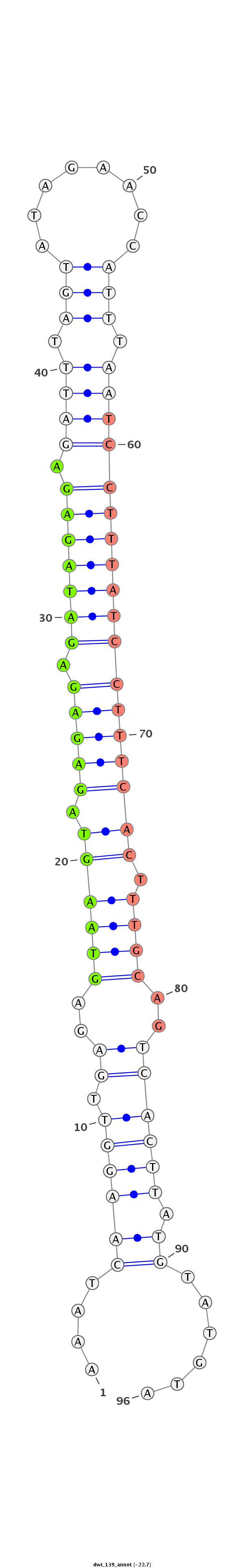
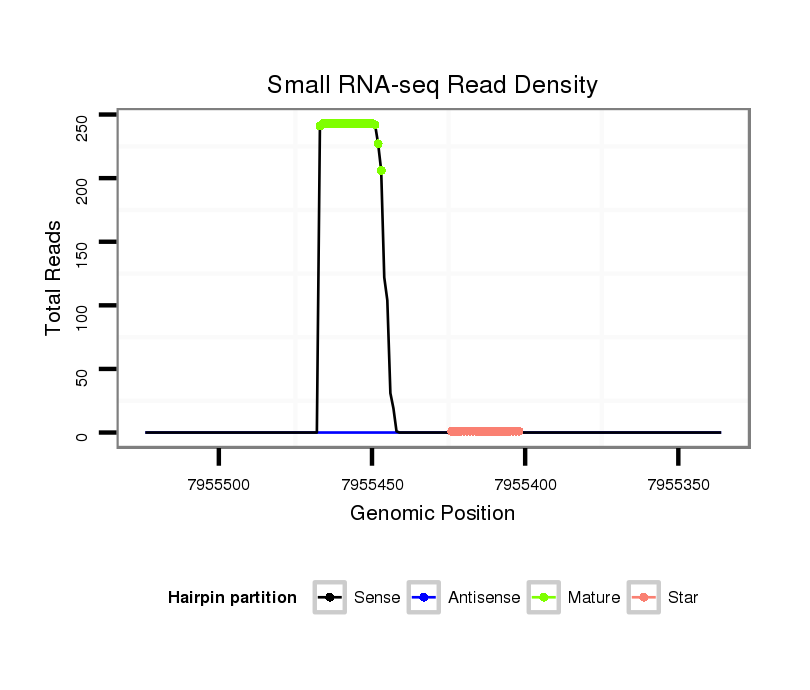
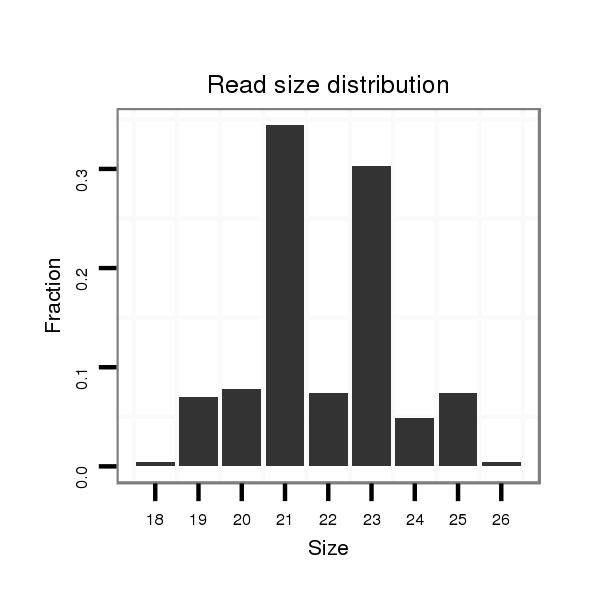
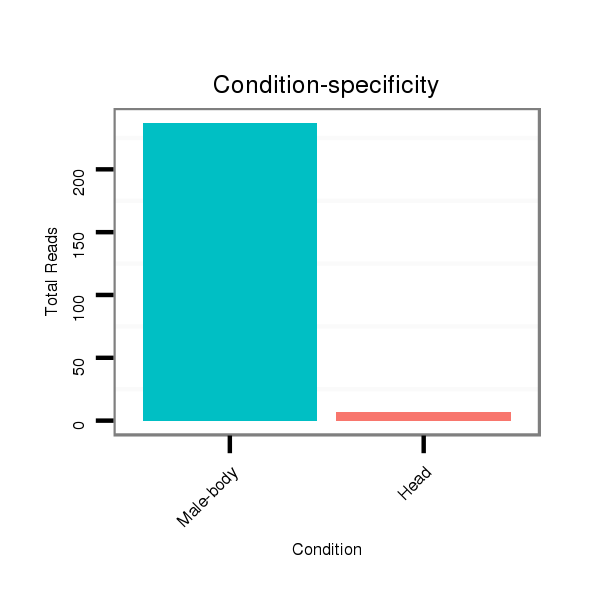
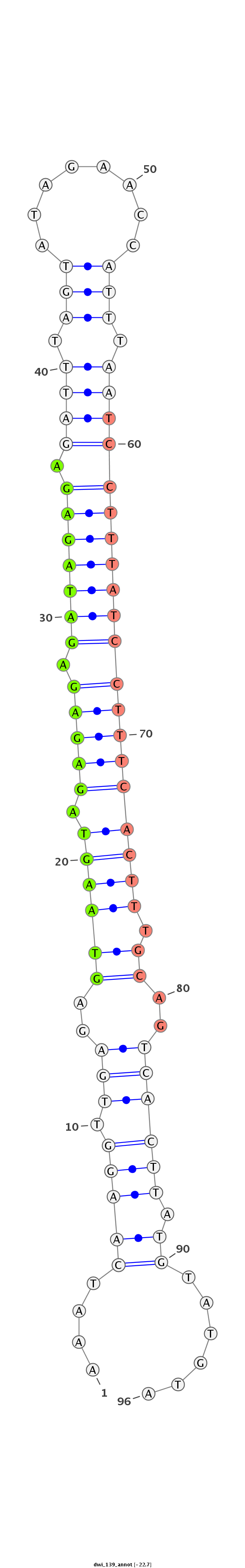
ID:dwi_139 |
Coordinate:scf2_1100000004909:7955386-7955474 - |
Confidence:confident |
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -22.7 | -22.7 | -22.7 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CACCATTCTCATAAAATTTCGTAGCAGCACAGCTTTAGAAGGAAATCAAGGTTGAGAGTAAGTAGAGAGAGATAGAGAGATTTAGTATAGAACCATTTAATCCTTTATCCTTTCACTTTGCAGTCACTTATGTATGTAACGGGACCAGTTTCTGCGATGGCGTTACAATGTTATTAAAGATACGATGTA
******************************************....((((((.((..((((((.(((((.(((((((.((((.(((........))).)))))))))))))))))).))))..)))))).))......*************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V117 male body |
V119 head |
M020 head |
V118 embryo |
M045 female body |
|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................GTAAGTAGAGAGAGATAGAGA............................................................................................................... | 21 | 0 | 1 | 84.00 | 84 | 82 | 2 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGA............................................................................................................. | 23 | 0 | 1 | 73.00 | 73 | 71 | 2 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAG................................................................................................................ | 20 | 0 | 1 | 19.00 | 19 | 19 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAG.............................................................................................................. | 22 | 0 | 1 | 18.00 | 18 | 18 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGATT........................................................................................................... | 25 | 0 | 1 | 18.00 | 18 | 18 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGA................................................................................................................. | 19 | 0 | 1 | 15.00 | 15 | 13 | 2 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGAT............................................................................................................ | 24 | 0 | 1 | 12.00 | 12 | 12 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAA.............................................................................................................. | 22 | 1 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 |
| ....................................................................................................TCCTTTATCCTTTCACTTTGCAGTT................................................................ | 25 | 1 | 1 | 3.00 | 3 | 0 | 1 | 2 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGT............................................................................................................. | 23 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGT............................................................................................................... | 21 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGATTG.......................................................................................................... | 26 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| ..........................................................TAAGTAGAGAGAGATAGAG................................................................................................................ | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGATC........................................................................................................... | 25 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGATTTTTT....................................................................................................... | 29 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGATTT.......................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGCAGAGAGAGATAGAGAGATT........................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATATAG................................................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGATTTC......................................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GGAAGTAGAGAGAGATAGAGAGATT........................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGATA........................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................TCCTTTATCCTTTCACTTTGCAG.................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGATTC.......................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................AGCACAGCTTTATGAGGAAA................................................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGATTTCC........................................................................................................ | 28 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATACAGA............................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGAA............................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGACAGA............................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAACTAGAGAGAGATAGAGAG.............................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAAAGATAGAGA............................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAG.................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGCGAGAGATAGAGAGA............................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAT.............................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAACTAGAGAGAGATAGAGAGATT........................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GTAAGTAGAGAGAGATAGAGAGTT............................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................AAAACCAGGTTGAGAGGAAGT.............................................................................................................................. | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...........................CACAGCTTTATGAGGAAATG.............................................................................................................................................. | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................AAAACCAGGTTGAGAGGAAG............................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ........................................................AGGAAGTAAAGAGAGATAG.................................................................................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
GTGGTAAGAGTATTTTAAAGCATCGTCGTGTCGAAATCTTCCTTTAGTTCCAACTCTCATTCATCTCTCTCTATCTCTCTAAATCATATCTTGGTAAATTAGGAAATAGGAAAGTGAAACGTCAGTGAATACATACATTGCCCTGGTCAAAGACGCTACCGCAATGTTACAATAATTTCTATGCTACAT
***************************************************....((((((.((..((((((.(((((.(((((((.((((.(((........))).)))))))))))))))))).))))..)))))).))......****************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V119 head |
M045 female body |
|---|---|---|---|---|---|---|---|
| .......................CGTCGCGTCGAAATTTTCCTG................................................................................................................................................. | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 |
| ...............................................................................................................................................TGGTCACAGACGCTATTGC........................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droWil2 | scf2_1100000004909:7955336-7955524 - | dwi_139 | confident | CACCATTCTCATAAAATTTCGTAGCAGCACAGCTTTAGAAGGAAATCAAGGTTGAGAGTAAGTAGAGAGAGATAGAGAGATTTAGTATAGAACCATTTAATCCTTTATCCTTTCACTTTGCAGTCACTTATGTATGTAACGGGACCAGTTTCTGCGATGGCGTTACAATGTTATTAAAGATACGATGTA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 10/20/2015 at 06:59 PM