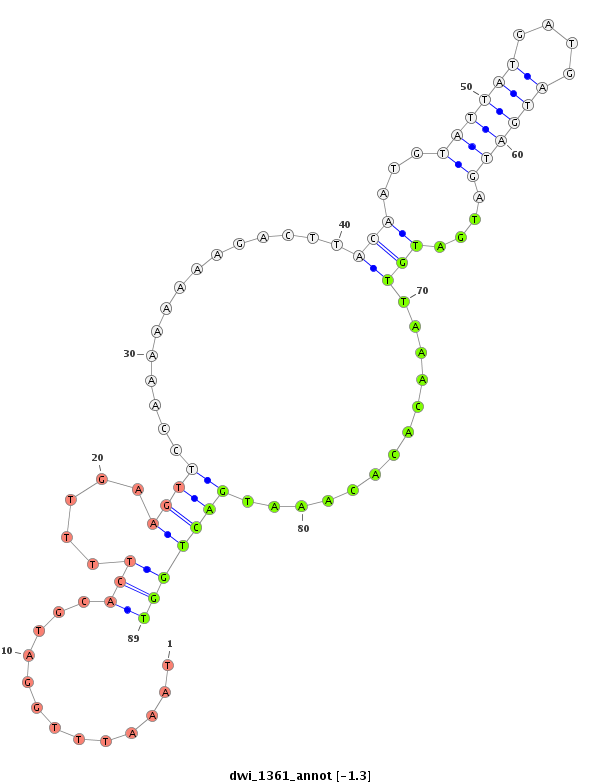
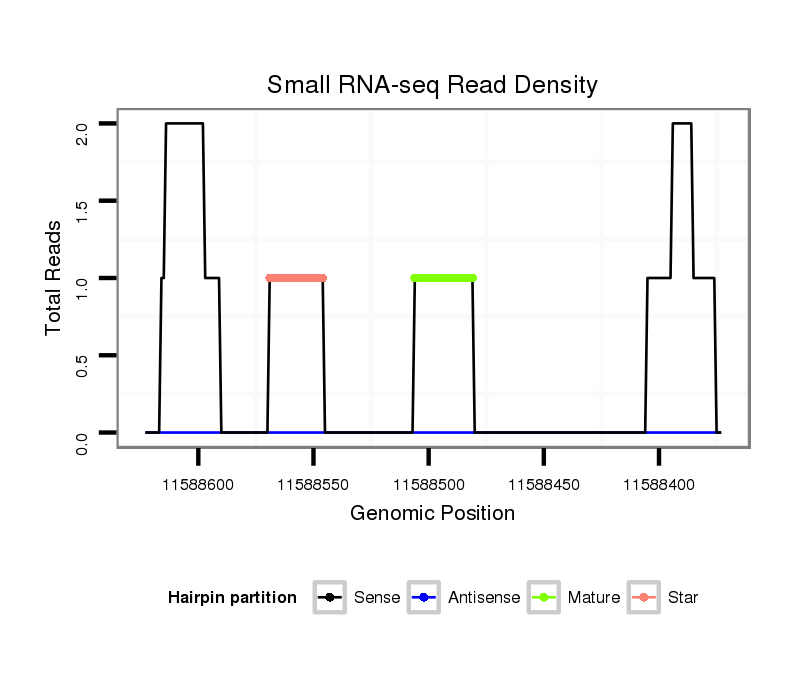
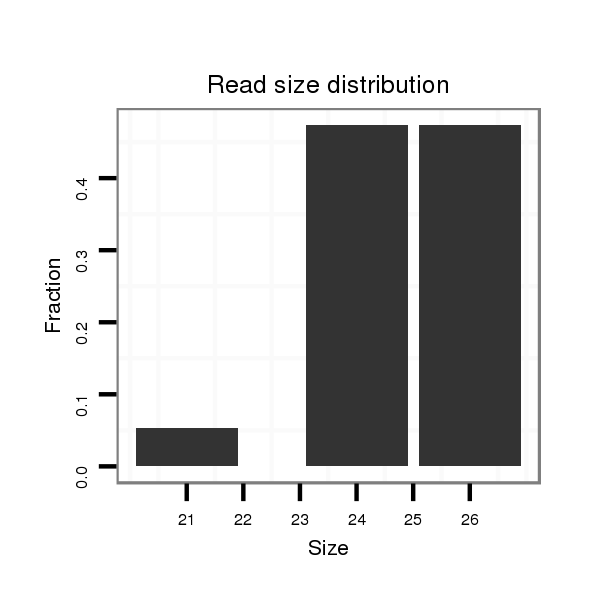
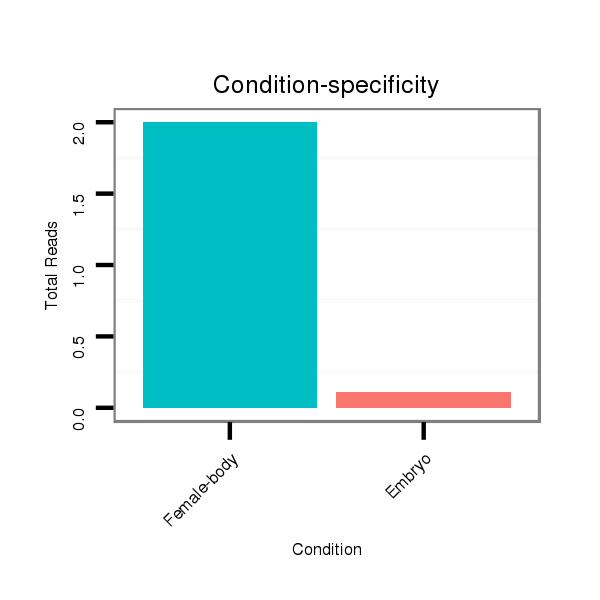
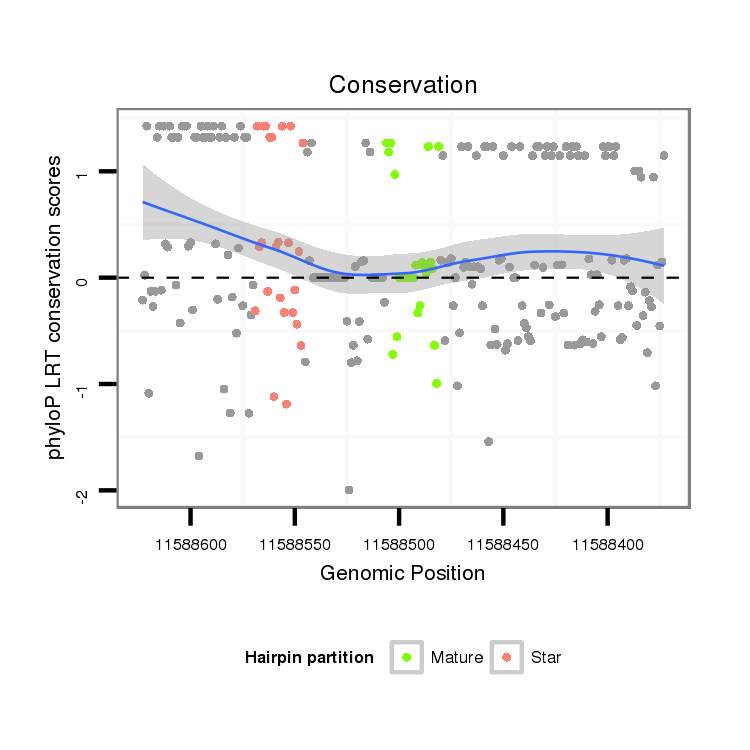
ID:dwi_1361 |
Coordinate:scf2_1100000004521:11588423-11588573 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -8.7 | -8.2 | -8.0 | -7.9 |
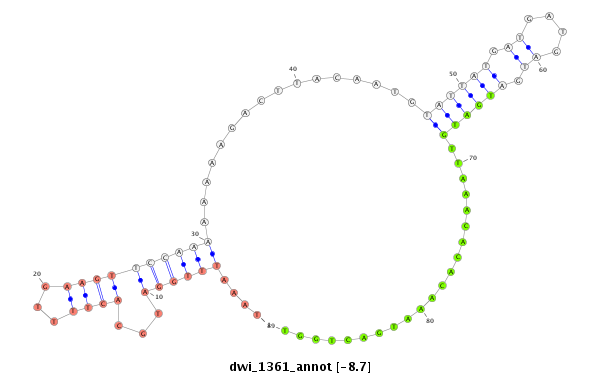 |
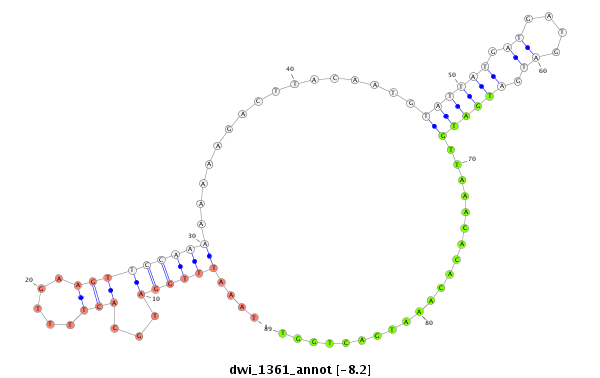 |
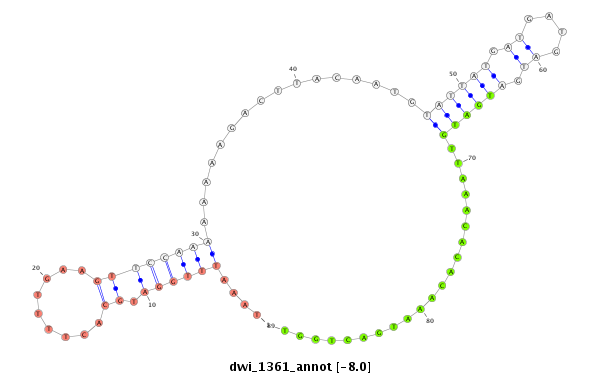 |
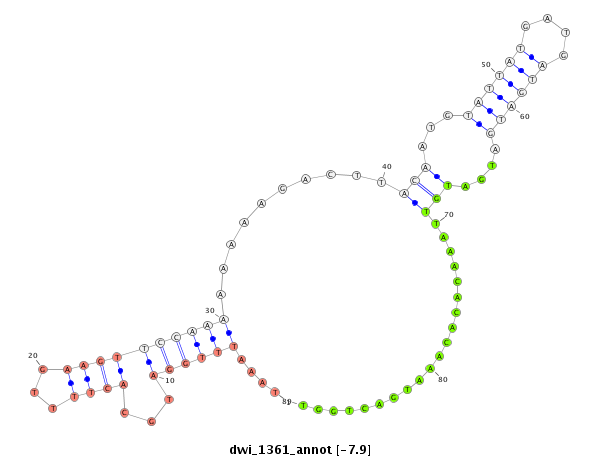 |
CDS [Dwil\GK14572-cds]; exon [dwil_GLEANR_14887:3]; intron [Dwil\GK14572-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------#####################----------------------------- CGATTGTCACTACAGCTGGAATGTCCCATGGTGGTATGTGGCCTCCATTGCCCATAAATTTGGATGCACTTTTGAAGTTCCAAAAAAAAGACTTACAATGTATTATGATGATGATGATGATGTTAAACACACAAATGACTGGTTGATTCTTCCACATTTTGATGATTCAACTGGCCAAAATAAACATCCATATGACCTTAGCATACTAACGGCCAGTTCTAAGTATGAACGAGATGTAAGCCCTCGGATGC ******************************************************.............(((.....((((...............(((...((((((....))))))....))).............)))))))************************************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
M045 female body |
V117 male body |
|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................................CGAGATGTAAGCCCTCGGA... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......CACTACAGCTGGAATGTCC................................................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....................................................................................................................TGATGTTAAACACACAAATGACTGGT............................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .........CTACAGCTGGAATGTCCCATGGTG.......................................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................TCTTCGACTCTTTGATGATTCA.................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................CTAAGTATGAACGAGATGTA............. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ......................................................TAAATTTGGATGCACTTTTGAAGT............................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .....................TGTCCAATGGCGGCATGTGGC................................................................................................................................................................................................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 |
| ...................................................................................................................................CAAATGGCTGGTTGTTTCTCC................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 |
| ........................................................................................................ATGATGATGATGATGATGTTA.............................................................................................................................. | 21 | 0 | 18 | 0.11 | 2 | 2 | 0 | 0 |
| .......................................................................................................................................TACCTGGTTGATTCTGCCA................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 1 | 1 | 0 |
|
GCTAACAGTGATGTCGACCTTACAGGGTACCACCATACACCGGAGGTAACGGGTATTTAAACCTACGTGAAAACTTCAAGGTTTTTTTTCTGAATGTTACATAATACTACTACTACTACTACAATTTGTGTGTTTACTGACCAACTAAGAAGGTGTAAAACTACTAAGTTGACCGGTTTTATTTGTAGGTATACTGGAATCGTATGATTGCCGGTCAAGATTCATACTTGCTCTACATTCGGGAGCCTACG
************************************************************************************************************.............(((.....((((...............(((...((((((....))))))....))).............)))))))****************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V119 head |
|---|---|---|---|---|---|---|---|
| ......................................................................................TTTCTGATTGTTAAATAATAC................................................................................................................................................ | 21 | 2 | 2 | 0.50 | 1 | 1 | 0 |
| ...................TTCCAGGATACCAGCATACA.................................................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 |
| ..................................................................GTGGAAACTTCTAGGTTT....................................................................................................................................................................... | 18 | 2 | 17 | 0.06 | 1 | 1 | 0 |
| ...................................................................TGAAAAGTCCAAGGTTTGTT.................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004521:11588373-11588623 - | dwi_1361 | CGATTGTCACTACAGCTGGAATGTCCCATGGTGGTATGTGGCCTCCATTGCCCATAAATTTGGATGCACTTTTGAAG----------------------------TTCCAAAAAAAAGACTTACAATGTATTATGATGATGATGATGAT--------GTTAAACACACAAATGACTGGTTGATTCTTCC---ACATT--T------TGATGATTCAACTGGCCAAAATAAACATCCATATGACCTTAGCATACTAACGGCCAGTTCTAAGTATGAACGAGA---TGTAAGCCCTCGGATGC |
| dp5 | 2:10050625-10050860 - | AGACACACACTACAGCTGGAATGTCCCTTGGTGGTACGTCGCTGCAGTCGCGGATAAATTCGGGCACAAGTTCACC---------------------------TGTGCCA----------------CGGACCCTGACG------GTGAGCTGATCATGG--------ATGAGGACTACTCGTTTTC-GGACAACATG--TTGGGTTGGCTAATAGAACCAGCTAA--AAGCACCCAATTCGATCTGAGTTTGCTGACATCATCCAGCCAGTACGAGAAGAG---GGTCAGCAAAAGCAAGC | |
| droPer2 | scaffold_0:10431248-10431483 + | AGACACACACTACAGCTGGAATGTCCCATGGTGGTACGTCGCTGCAGTCGCGGATAAATTCGGGCACAAGTTCACC---------------------------TGTGCCA----------------CAGACCCTGACG------GTGAGCTGATCATGG--------ATGAGGACTACTCGTTTTC-GGACAACATG--TTGGGTTGGCTAATTGAACCAGCTAA--AAGCACCCAATTCGATCTGAGTTTGCTGACATCATCCAGCCAGTACGAGAAGAG---GGTCAGCAAAAGCAAGC | |
| droAna3 | scaffold_12943:2341743-2341966 - | TGACGTGCATTACAGCTGGAATACACCATGGTGGTACGTTGCAGCAATAGCAGAAAAGTTCGGCCACAAATTTGTAT---------------------------GTTCTA----------------CACCTGCCAATG------GTGAACTAATATTGG--------ATGGGGATTATTCGGAATC-TAACAACTTG--ATCGGATGGCTAATAGAGTCAGGCCA--CAGTGCGCAATTTGACCTAAGCTTACTAACGTCATCGTGTCAGTACGAGACGAG---------------AAAAC | |
| droKik1 | scf7180000301798:455-663 - | AAAGACACACTTTAGCGGTAATGTTCCTTGGTGGTTTGTTGGGGCTATTGCTGCCACATTTGGGCAAAACTTCAAAG---------------------------------------------------------------------------------T--------AAAAGCACGCGTCGATTTT-AAACAAAATGAAATAGGATGGCGGATAAATGCAGAAAA--TAACGAGCAATATGATCTAGGCATTCTTACTGCGTCCAGCAAGTACGAACAGAAAGTGTTAAGGAGTCGTGGGC | |
| droTak1 | scf7180000413017:216956-217082 + | AAAAACACATTACAGCGGCAATGTTCCTTGGTGGTATGTGGCCACTATTGCCTCTAAATTCGGTCATACTATTAAAATAGAGGCATCTTCCCAAGAAGATTATAGTCCCA----------------ATGGTTTTGAAG------ATGAT-------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/18/2015 at 01:17 AM