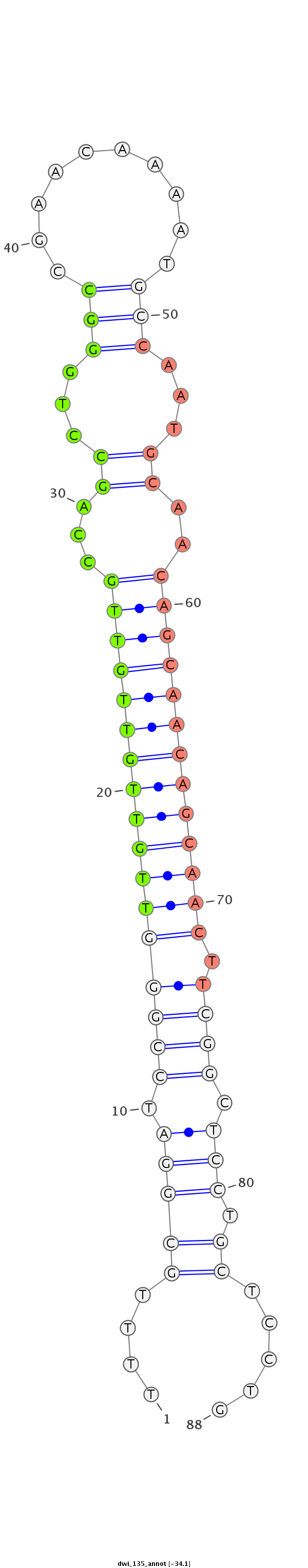
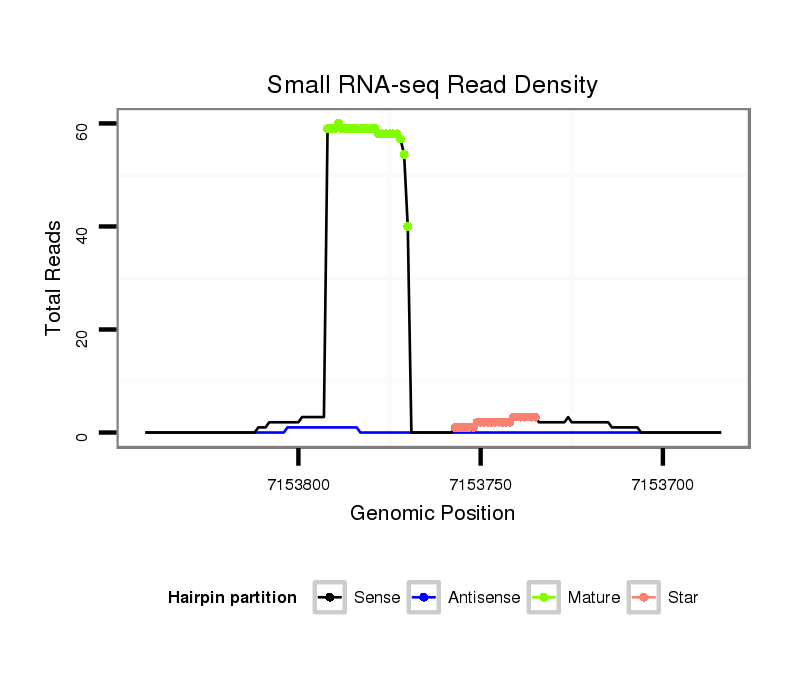
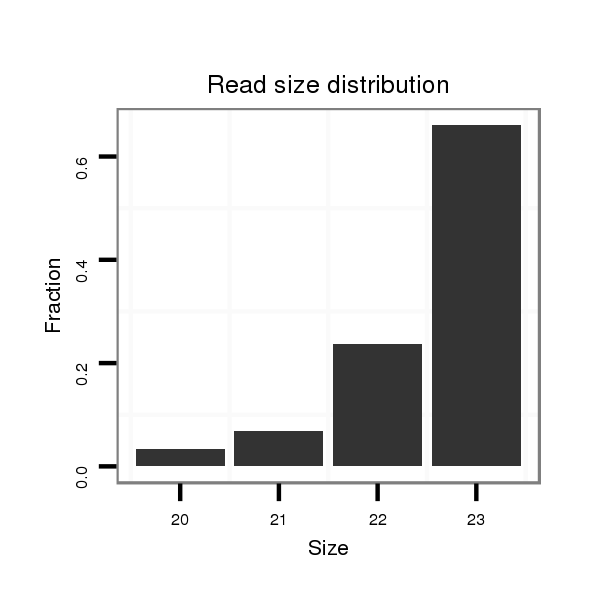
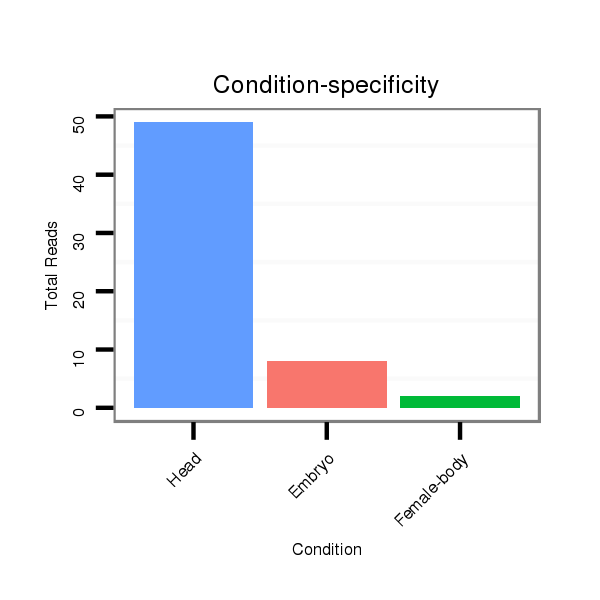
ID:dwi_135 |
Coordinate:scf2_1100000004909:7153734-7153792 - |
Confidence:candidate |
Class:Canonical miRNA |
Genomic Locale:CDS |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -34.1 |
 |
CDS [Dwil\GK25170-cds]
No Repeatable elements found
|
CAATTGACATGAAGACAATTTTACTTTTGTGGATGTTTTGCGGATCCGGGTTGTTGTTGTTGCCAGCCTGGGCCGAACAAAATGCCAATGCAACAGCAACAGCAACTTCGGCTCCTGCTCCTGCTTCAGCTGCGGCTGCCGCTTCCAGCAATCCTGCCC
***********************************....(((((.(((((((((((((((((...((...(((..........)))...))..))))))))))))).)))).))).)).....************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V119 head |
V118 embryo |
M045 female body |
V117 male body |
M020 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TTGTTGTTGTTGCCAGCCTGGGC...................................................................................... | 23 | 0 | 1 | 38.00 | 38 | 33 | 5 | 0 | 0 | 0 |
| ..................................................TTGTTGTTGTTGCCAGCCTGGG....................................................................................... | 22 | 0 | 1 | 14.00 | 14 | 11 | 2 | 0 | 0 | 1 |
| ..................................................TTGTTGTTGTTGCCAGCCTGG........................................................................................ | 21 | 0 | 1 | 3.00 | 3 | 2 | 0 | 1 | 0 | 0 |
| ...........................................ATCCGGGTTGTTGTTGTTGCC............................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TTGTTGTTGTTGCCAGCCTAGGC...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TTGTTGTTGTTGACAGCCTGGGC...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................GTTGTTGTTGTTGCCATCCTGGGCC..................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TTGTTGTTGTTGCCAGCCTG......................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TTGTTGTTGTTGCCAGCCTGGC....................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................GCTCCTGCTTCAGCTGCGGC....................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................CAATGCAACAGCAACAGCAACTT................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................TTGTTGTTGCCAGCCTGGGC...................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................GTTGTTGTTGCCAGCCTGGGC...................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TTGTTGTTGTTGCCAGTCTGGG....................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................AACAGCAACAGCAACTTCGGCTCCTG.......................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................GCAACTTCGGCTCCTGCTCCTGCTTCA............................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TTGTTGTTGTTGCAAGCCTGGGC...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TTGTTGTTGTTGCCAGCCTGGAC...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................GTTTTGCGGATCCGGGTT........................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TTGTTGTTGTTGCCAGCCTGGGT...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TTGTTGTTGTTGCCAGCCTGGGCCGT................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TTGTTGTTGTTTCCAGCCTGGG....................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................GATGTTTTGCGGATCCGGGTTGT......................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................TTCTTGTTGTTGCCAGCCTGGGC...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................TTTGGGGATGTTTTGCGGATCCGGGTT........................................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................TTGTTGTTGTTGCCACCCTGGGC...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................GGCGGAACCGGGTTGGTGTT..................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................GATCCGGGTTGAAGTCGTT.................................................................................................. | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 |
|
GTTAACTGTACTTCTGTTAAAATGAAAACACCTACAAAACGCCTAGGCCCAACAACAACAACGGTCGGACCCGGCTTGTTTTACGGTTACGTTGTCGTTGTCGTTGAAGCCGAGGACGAGGACGAAGTCGACGCCGACGGCGAAGGTCGTTAGGACGGG
************************************....(((((.(((((((((((((((((...((...(((..........)))...))..))))))))))))).)))).))).)).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
M045 female body |
V117 male body |
|---|---|---|---|---|---|---|---|---|
| .......................................CGCCTAGGCCCAACAACAAC.................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........................................................................................................................................GCGAAGGACGTTGGGACG.. | 18 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ..................................CAAAACGACTAGGCGCCACA......................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 |
| ......................................................................................................................................CGACGGAGAAGGTCGGTA....... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 |
| ................................................................................................................AGGACTAGGGCGAAGTCG............................. | 18 | 2 | 17 | 0.06 | 1 | 0 | 0 | 1 |
| .................................................................................................TAGGCGTCGAAGCCGAGGA........................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droWil2 | scf2_1100000004909:7153684-7153842 - | dwi_135 | candidate | CAATTGACATGAAGACAATTTTACTTTTGTGGATGTTTTGCGGATCCGGGTTGTTGTTGTTGCCAGCCTGGGCCGAACAAAATGCCAATGCAACAGCAACAGCAACTTCGGCTCCTGCTCCTGCTTCAGCTGCGGCTGCCGCTTCCAGCAATCCTGCCC |
| droGri2 | scaffold_15203:3522820-3522879 + | --------------------------------------------------------------------------------------AATGCAACAACGACAGCCACAGCCGTTTCGGCTGTGTCTGCAGCAGCAGCTGCAGCGTCC------------- | ||
| droPer2 | scaffold_13:308965-309012 + | --------------------------------------------------------------------------------------------------------------GCTCCTGCTGCTGCTGCTGCTGCTGCTGACGCGTTTCGTGGCTGTGCC- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 10/20/2015 at 07:44 PM