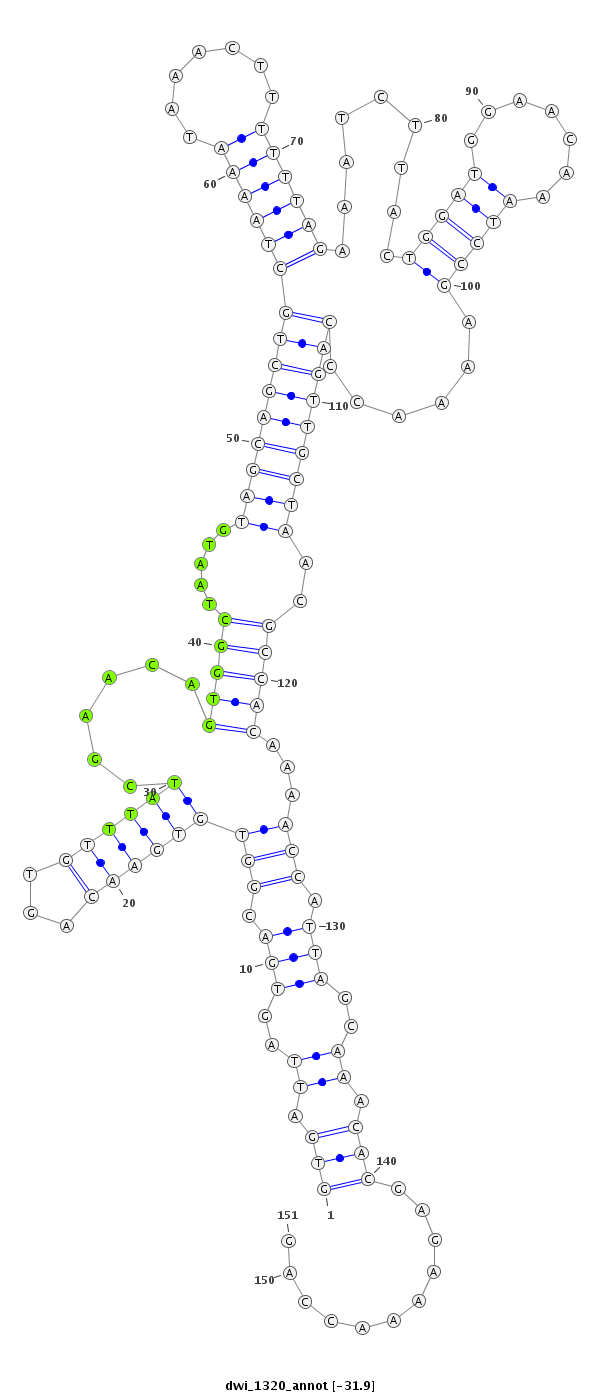
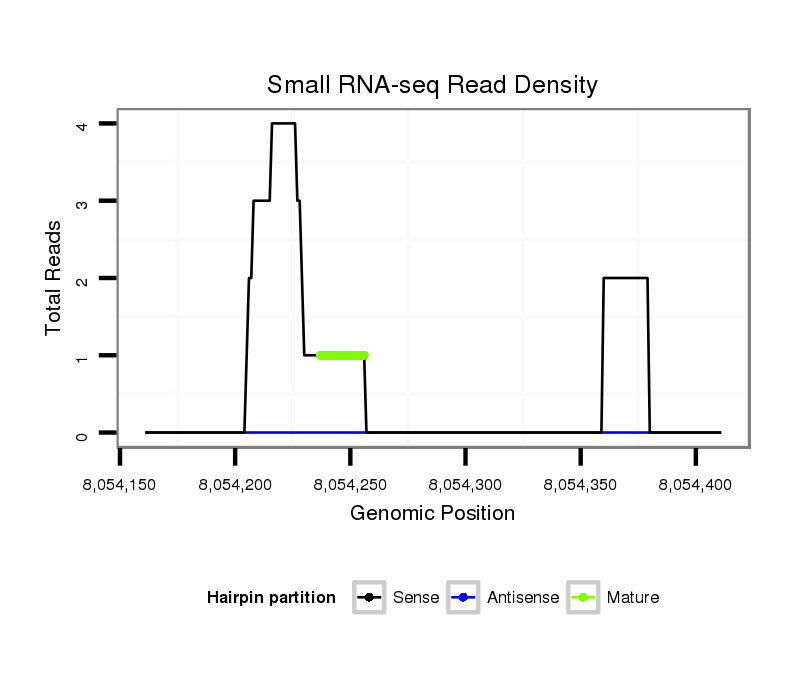
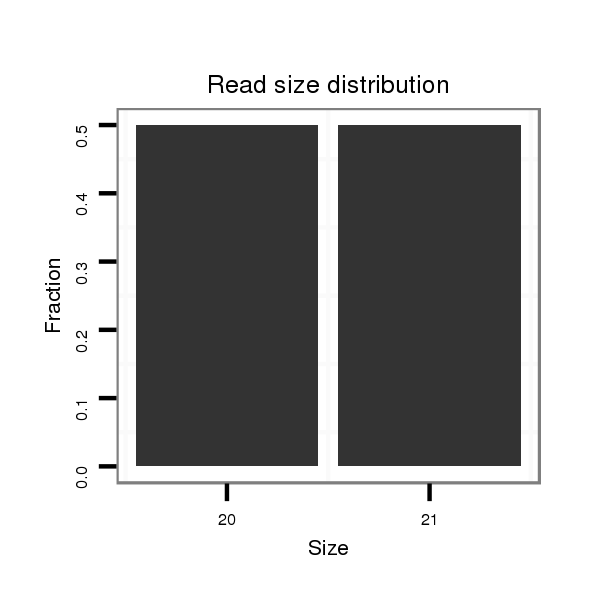
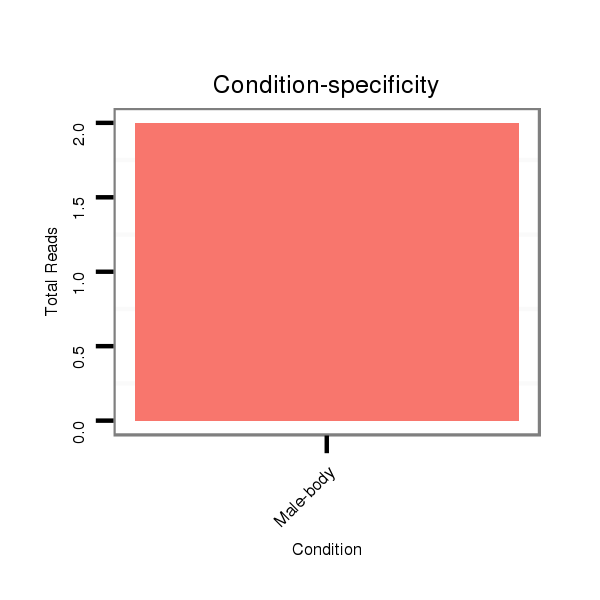
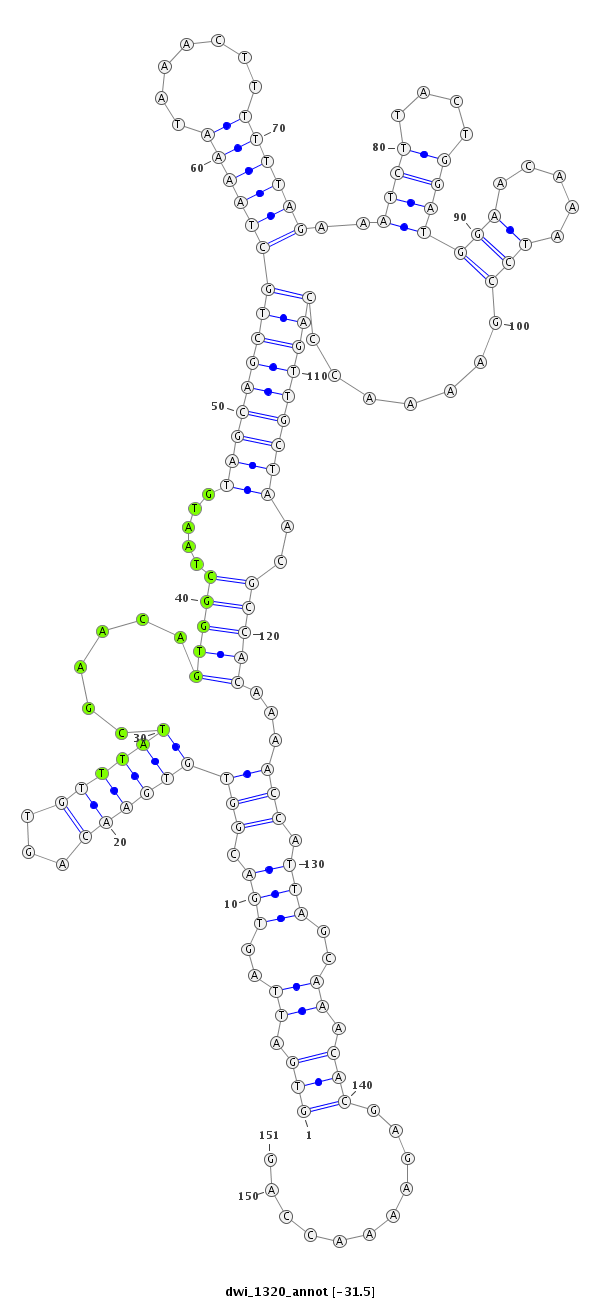
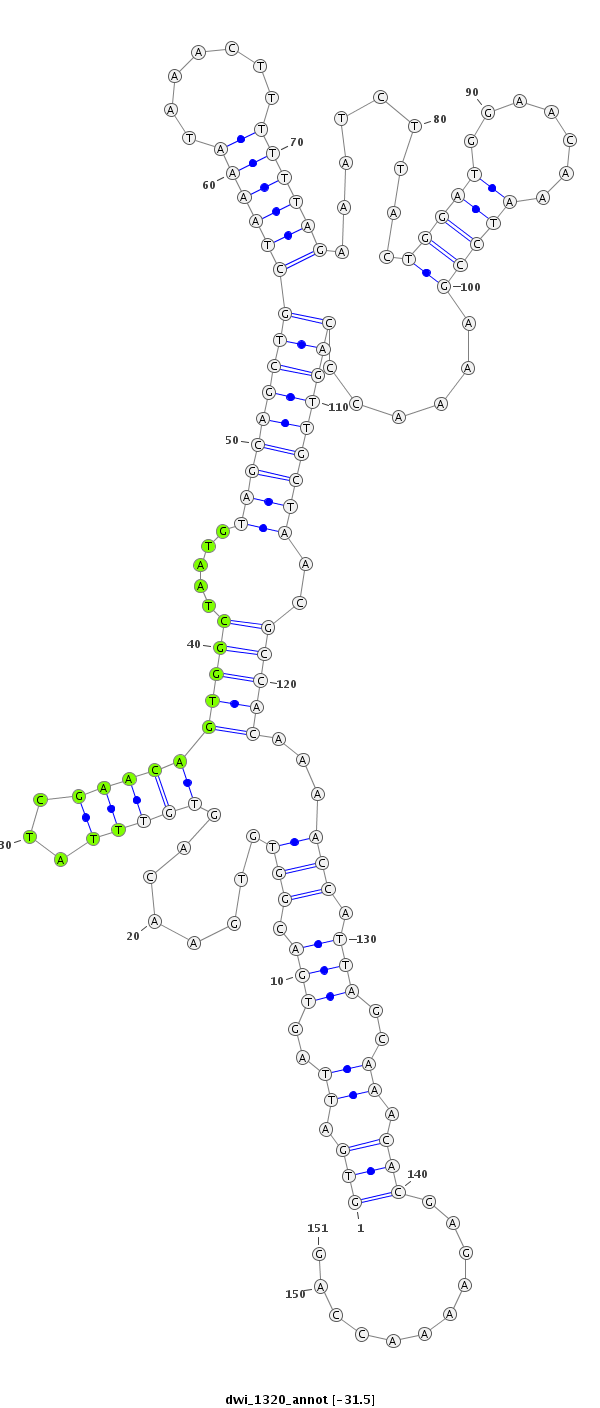
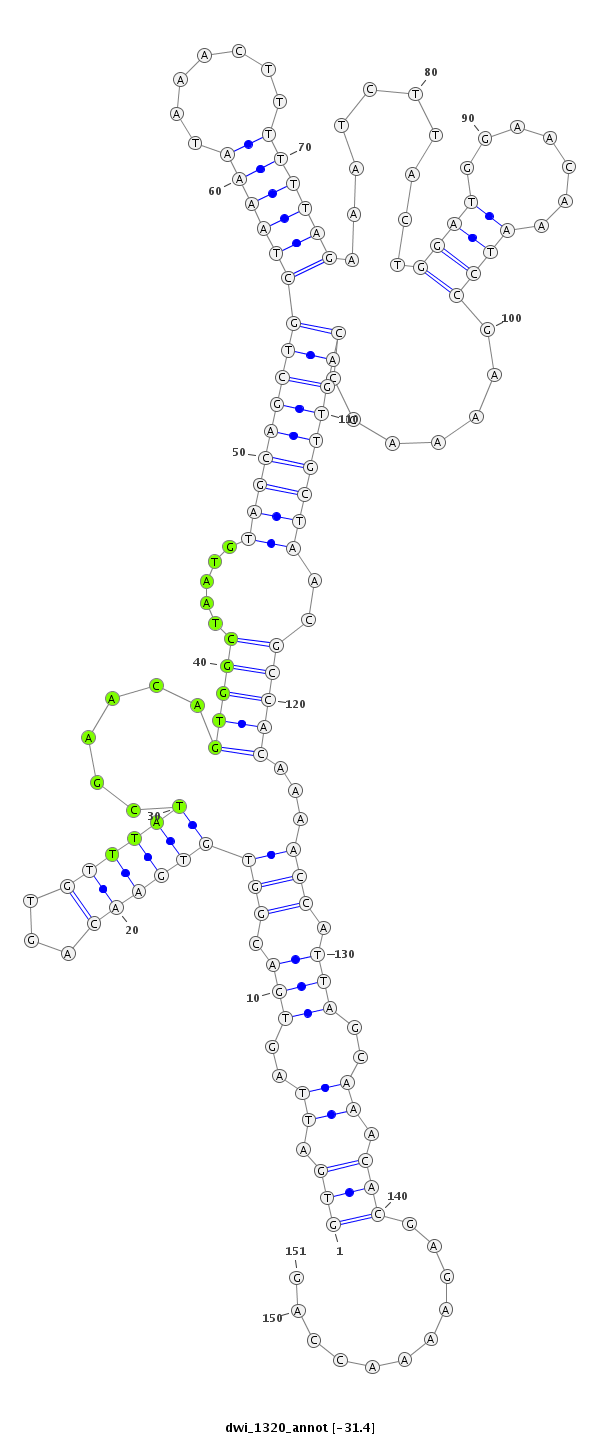
ID:dwi_1320 |
Coordinate:scf2_1100000004521:8054211-8054361 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -31.5 | -31.5 | -31.4 |
 |
 |
 |
CDS [Dwil\GK15413-cds]; exon [dwil_GLEANR_15666:2]; intron [Dwil\GK15413-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATGCTGCCTCCAACAAACCACCTGCCATTTCCAAAAGAACTGTCACTAAAGTGATTAGTGACGGTGTGAACAGTGTTTATCGAACAGTGGCTAATGTAGCAGCTGCTAAAATAAACTTTTTTAGAAATCTTACTGGATGGAACAAATCCGAAAACCCAGTTGCTAACGCCACAAAACCATTAGCAAACACGAGAAAACCAGTTAATTATTCGCAAAGACCAGTTACTTACTTAGAGGAACAACATACTTAC **************************************************(((.((..(((.(((((((((...))))))......(((((.....(((((((((((((((.......)))))).........(((((.......)))))......)))))))))..)))))...))).)))..)).)))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V119 head |
V117 male body |
M020 head |
V118 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ..............................................................CGTGTGAACAGTGTTGATC.......................................................................................................................................................................... | 19 | 2 | 1 | 5.00 | 5 | 4 | 1 | 0 | 0 |
| ..............................................................CGTGTGAACAGTGGTTATC.......................................................................................................................................................................... | 19 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ..............................................................CGTGTGAACAGTGTTGAT........................................................................................................................................................................... | 18 | 2 | 3 | 2.00 | 6 | 6 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................AGTTAATTATTCGCAAAGAC................................ | 20 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 |
| ....................................................................................................................................................................................................ACCAGTTAATGATTCGCGAT................................... | 20 | 3 | 5 | 1.20 | 6 | 0 | 0 | 6 | 0 |
| ............................................ACTAAAGTGATTAGTGACGGTGTGA...................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................GTGTGAACAGTGTTGATC.......................................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................TTATCGAACAGTGGCTAATG........................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................CTAAAGTGATTAGTGACGGTG......................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .......................................................TAGTGACGGTGTGAACAGTGT............................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...............................................AAAGTGATTAGTGACGGTGTG....................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................................TCGTGTGAACAGTGTTGATC.......................................................................................................................................................................... | 20 | 3 | 4 | 0.75 | 3 | 3 | 0 | 0 | 0 |
| ...............................................................GTGTGAACAGTGGTAATC.......................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...........AACAAACCGGCTGGCATTTCC........................................................................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................CGTGTGAACAGTGTTGATCA......................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................CGTGTGAACAGGGTTGATC.......................................................................................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| .....................................................ATTTGTGACCGTGCGAACA................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .....................................................................................AGTGGCTGATGGAGCAGC.................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
TACGACGGAGGTTGTTTGGTGGACGGTAAAGGTTTTCTTGACAGTGATTTCACTAATCACTGCCACACTTGTCACAAATAGCTTGTCACCGATTACATCGTCGACGATTTTATTTGAAAAAATCTTTAGAATGACCTACCTTGTTTAGGCTTTTGGGTCAACGATTGCGGTGTTTTGGTAATCGTTTGTGCTCTTTTGGTCAATTAATAAGCGTTTCTGGTCAATGAATGAATCTCCTTGTTGTATGAATG
**************************************************(((.((..(((.(((((((((...))))))......(((((.....(((((((((((((((.......)))))).........(((((.......)))))......)))))))))..)))))...))).)))..)).)))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
M045 female body |
|---|---|---|---|---|---|---|---|
| ..........GTTGTGTGGTAGACGGTA............................................................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 |
| ............................................................................................................................................................................TTTTGGTGATCGTTGGGGCT........................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004521:8054161-8054411 + | dwi_1320 | ATGCTGCCTCCAACAAACCACCTGCCATTTCCAAAAGAACTGTCACTAAAGTGATTAGTGACGGTGTGAACAGTGTTTATCGAACAGTGGCTAATGTAGCAGCTGCTAAAATAAACTTTTTTAGAAATCTTACTGGATGGAACAAATCCGAAAACCCAGTTGCTAACGCCACAAAACCATTAGCAAACACGAGAAAACCAGTTAATTATTCGCAAAGACCAGTTACTTACTTAGAGGAACAACATACTTAC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 05/18/2015 at 01:11 AM