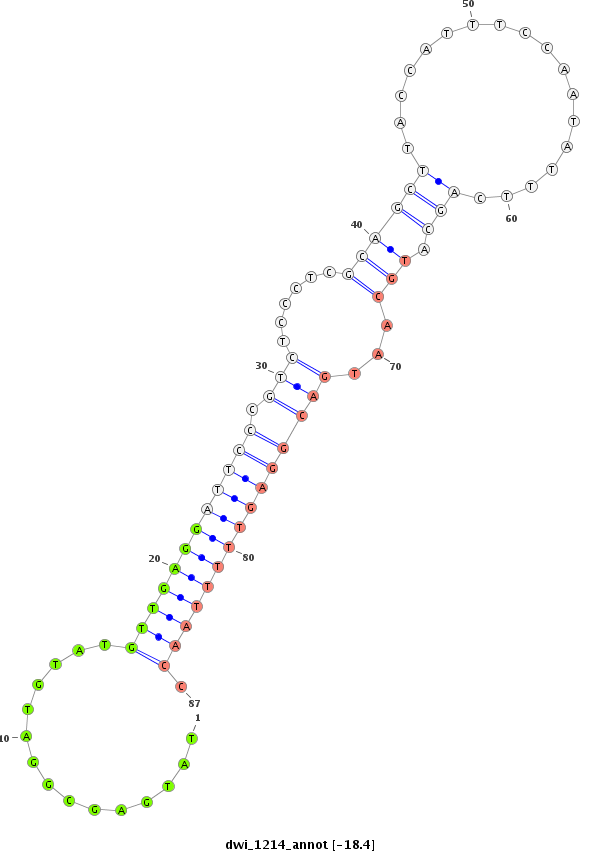
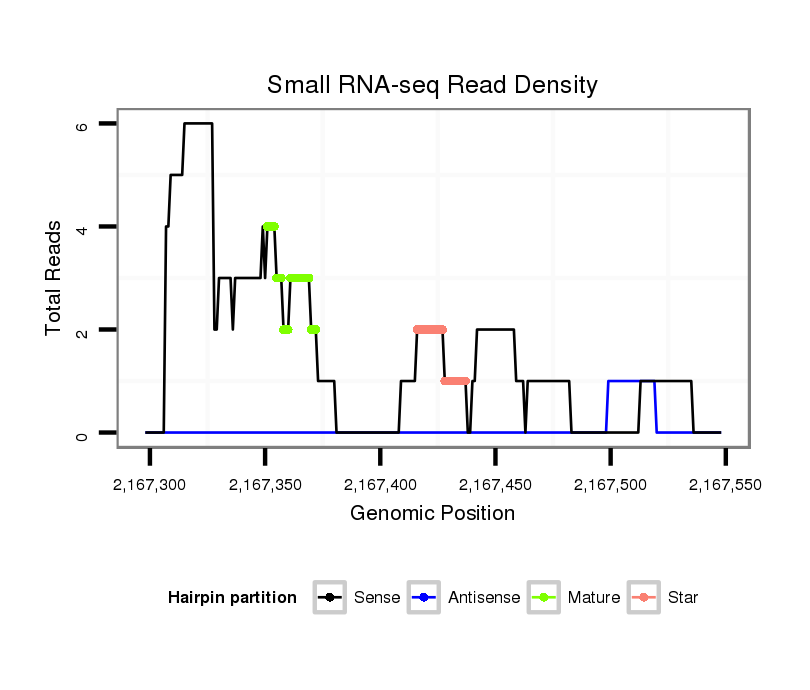
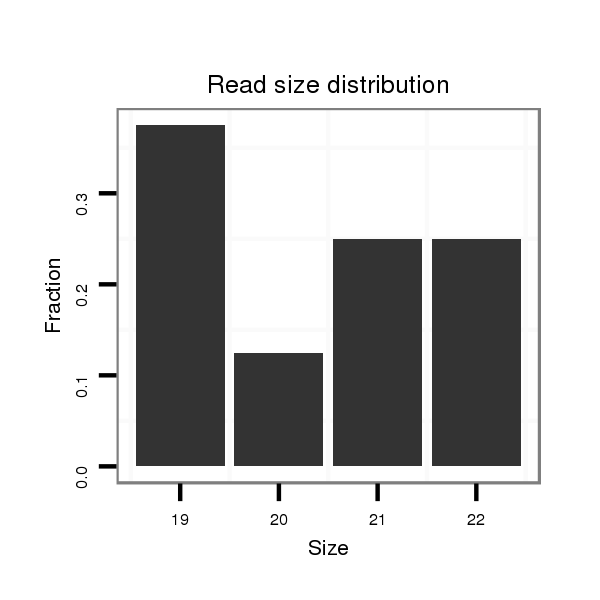
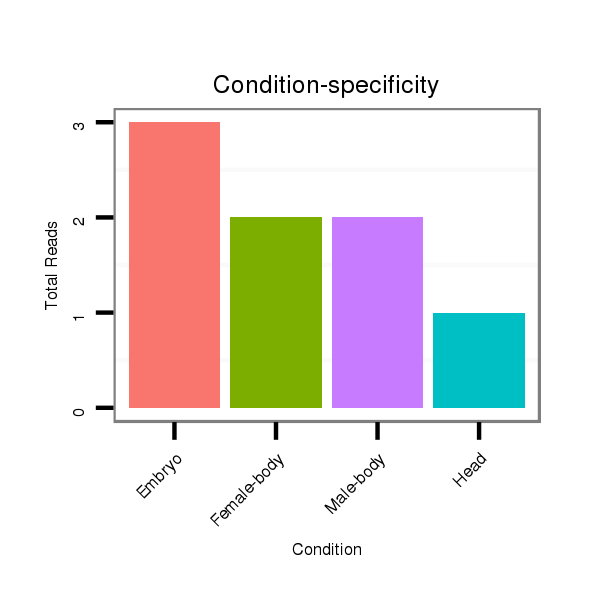
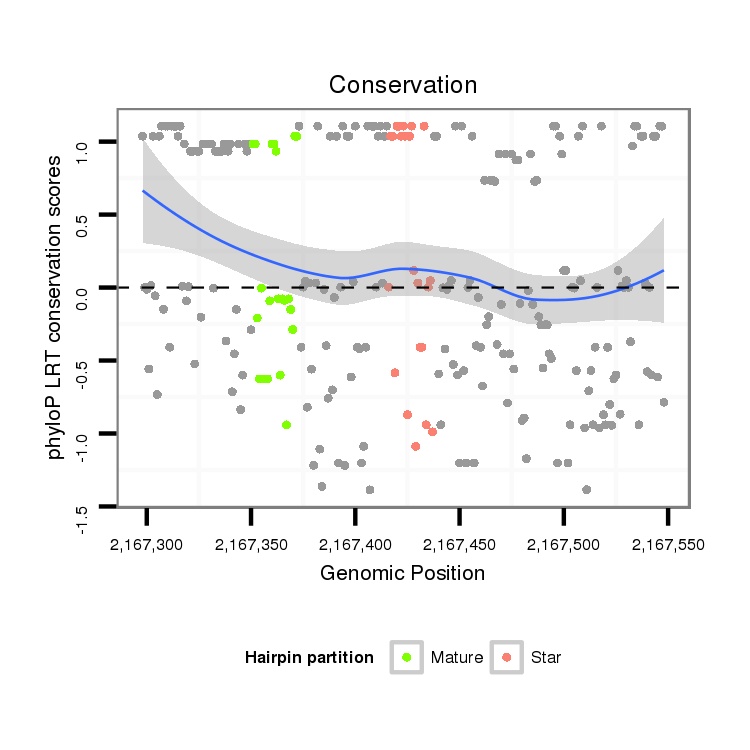
ID:dwi_1214 |
Coordinate:scf2_1100000004521:2167348-2167498 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -18.4 | -18.4 |
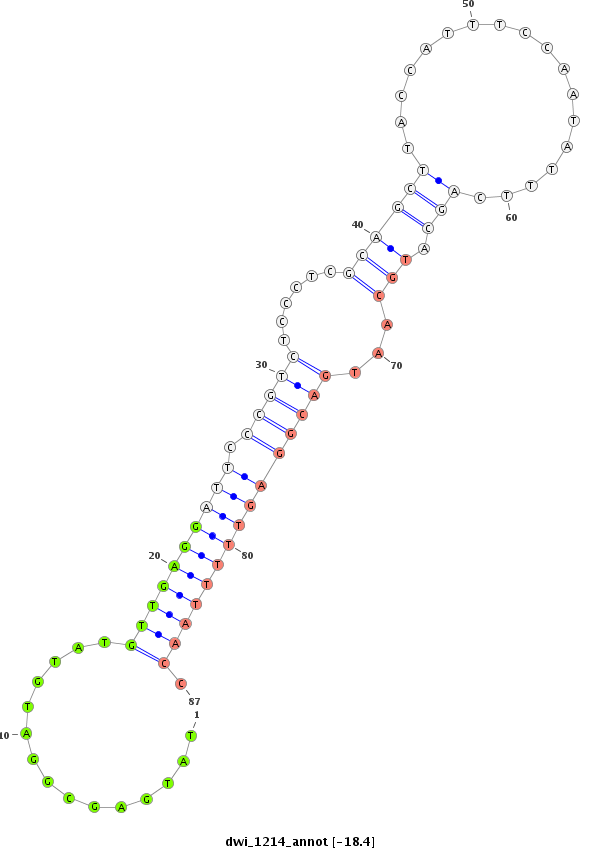 |
 |
CDS [Dwil\GK15144-cds]; exon [dwil_GLEANR_15417:6]; intron [Dwil\GK15144-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CACTTGCGCAAAATTTTGATTGCGCTGGGTTTATACCGCTAGTTCGACAAGTATATGAGCGGATGTATGTTGAGGATTCCCGTCTCCCTCGCAGCTTACCATTTCCAATATTTCAGCATGCAATGACGGAGTTTTTAACCGCGTATATGATACATCAGGGTAAATTTATCTTAAAGATTCCGGCTTTACAATCTTTAATGGACCCATTAGCAGCCATTAGTGCTGAGAGCTACAATATTCCAAAACCTATA *****************************************************...............((((((((((((.(((......((((((..................))).)))...))))))))))))))).*************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
V117 male body |
M045 female body |
V119 head |
M020 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .........AAAATTTTGATTGCGCTGGGT............................................................................................................................................................................................................................. | 21 | 0 | 1 | 3.00 | 3 | 1 | 2 | 0 | 0 | 0 |
| .....................................................TATGAGCGGATGTATGTTGAGG................................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................TGCAATGACGGAGTTTTTAACC............................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................ATACCGCTAGTTCGACAAGT....................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................TGTATGTTGAGGATTCCCGT........................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........AATTTTGATTGCGCTGGGT............................................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................TATATGAGCGGATGTATGTTG................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................TATACCGCTAGTTCGACAAGTATATG.................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................ATTAGTGCTGAGAGCTACAATAT............. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................GATTGCGCTGGGTTTATACCG..................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........AAAATTTTGATTGCGCTGGGTT............................................................................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................TAGTTCGACAAGTATATGAGC............................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GTATATGATACATCAGGGT.......................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................TATCTTAAAGATTCCGGCT.................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................ATATGATACATCAGGGTAAAT...................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................TTCAGCATGCAATGACGGA......................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CAGGGTAACTATAACTTAAAG........................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................TGGGGATTCCGGCTTTACAAT........................................................... | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................ATCTGAGCGGATGTGTGGT.................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................ACGGATGAATGTTGAAGAT.............................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
|
GTGAACGCGTTTTAAAACTAACGCGACCCAAATATGGCGATCAAGCTGTTCATATACTCGCCTACATACAACTCCTAAGGGCAGAGGGAGCGTCGAATGGTAAAGGTTATAAAGTCGTACGTTACTGCCTCAAAAATTGGCGCATATACTATGTAGTCCCATTTAAATAGAATTTCTAAGGCCGAAATGTTAGAAATTACCTGGGTAATCGTCGGTAATCACGACTCTCGATGTTATAAGGTTTTGGATAT
***************************************************************************************************************...............((((((((((((.(((......((((((..................))).)))...))))))))))))))).***************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................TATGTGGTCCCAGTTAAATAGCAT.............................................................................. | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 |
| .........................................................................CCTAAGGGGAGAGGGCGCGT.............................................................................................................................................................. | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| .........................................................................................................................................................................................................TGGGTAATCGTCGGTAATCAC............................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ..........................................................................................CGTTGCATCGTAAAGGTTATAA........................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 |
| .........................................................................CCTAGGGGCAGATGGAGTGT.............................................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 |
| ...........................................................................................................................................................GTCCCAGTTAAATAGCAT.............................................................................. | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004521:2167298-2167548 + | dwi_1214 | CACT--TGCGCAAAATTTTGATTGCGCTGGGTTTATACCGCTAGTTCGACAAGTATATGAGCGGATGTATGTTGAGGATTCCCGTCTCCCTCGCAGCTTACCATTTCCAATATTTCAGCATGCAATGACGGAGTTTTTAACCGCGTATATGATACATCAGGGTAAATTTATCTTAAAGATTCCGGCTTTACAATCTTTAATGGACCCATTAGCAGCCATTAGTGCTGAGAGCTACAATATTCCAAAACCTATA |
| droKik1 | scf7180000302270:261881-262096 + | CT----TGGCCAAAACTTTGACTTTGCTGGCTTTATTCCGCTTGTAAAATCAGTATATATATATATGGCGGCGAGGGACTACCACCTCGAGAGACGGTTGCCCTTCTGTGTCTTTCAACATGCCATGACTGAAATCCTTACGGCAAGAATGTTGGAAGTGGCGCGG------C--------------------------ATCG-TTGTTATGCAAAATACTATTACGAAGATTTCTTTATACCACTGCCGATC | |
| droEle1 | scf7180000491024:259447-259642 - | CATA--AGGCCAGAATTTTGA------------------------------------------------------GGATCAAAAGATGACATGAAGCCTCCCTTTTCCAATTTTCCAGCACGCGATGACGGAATTTTTAGTTGCCTACCAATTGTATCAAGGAAATTACATCCTCAATATACCTGAGCTCCAGAGAATGATGGATTCAC-AGTAGCAATTAGTGCTAGTGAGTATAATATTCCACAGCCAATA | |
| droTak1 | scf7180000414050:285745-285982 + | CATTTGTGGTCAGAACTTTGACTTTGCGGGCTTTATTCCGCTAGTCCGAGAAGTTTACTTTATTATGGAGAGGGAGGACTATCTTCTACCAAGGAGGTTACCCATCTGCGTGTTTCAGCATGCGATGACTGAAGCCCTGACGGCCCACATGATTCAAGAGGCAAGG------TTCGAAGTTCCTCGCCTC-------CAATCGATTGGTATGCATGTTCCATTTTCTTATATGACA--ATCCCGTATCCGATC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/18/2015 at 12:55 AM