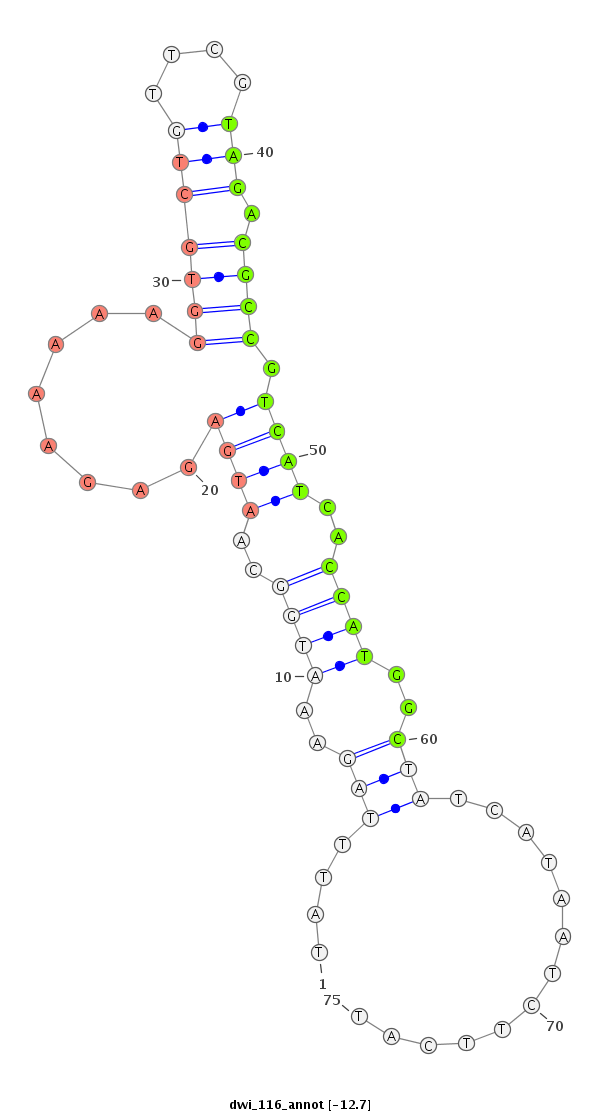
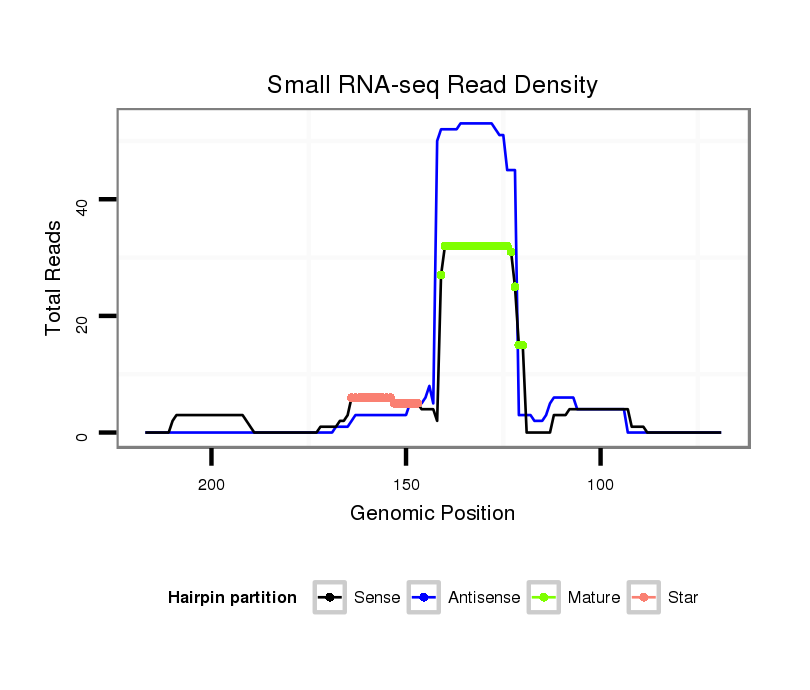
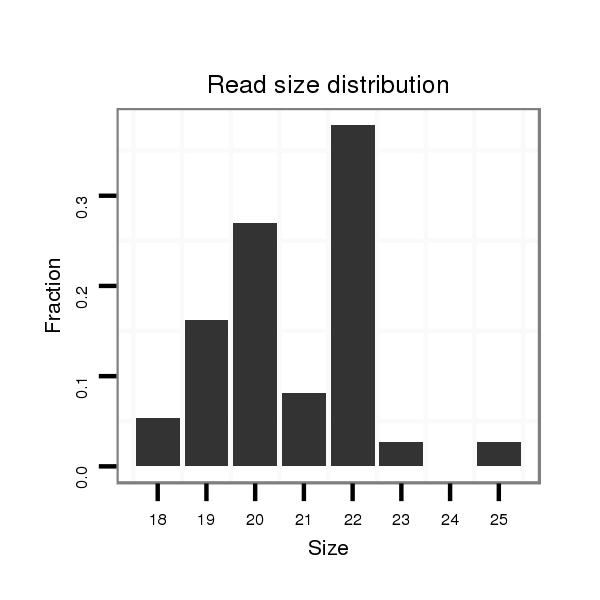
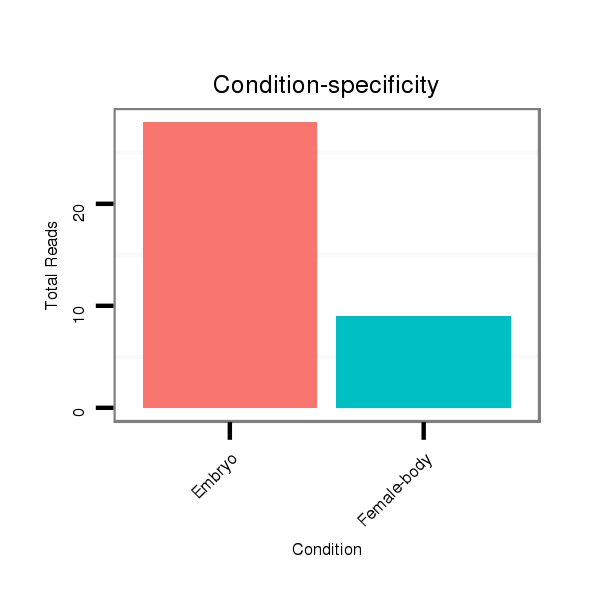
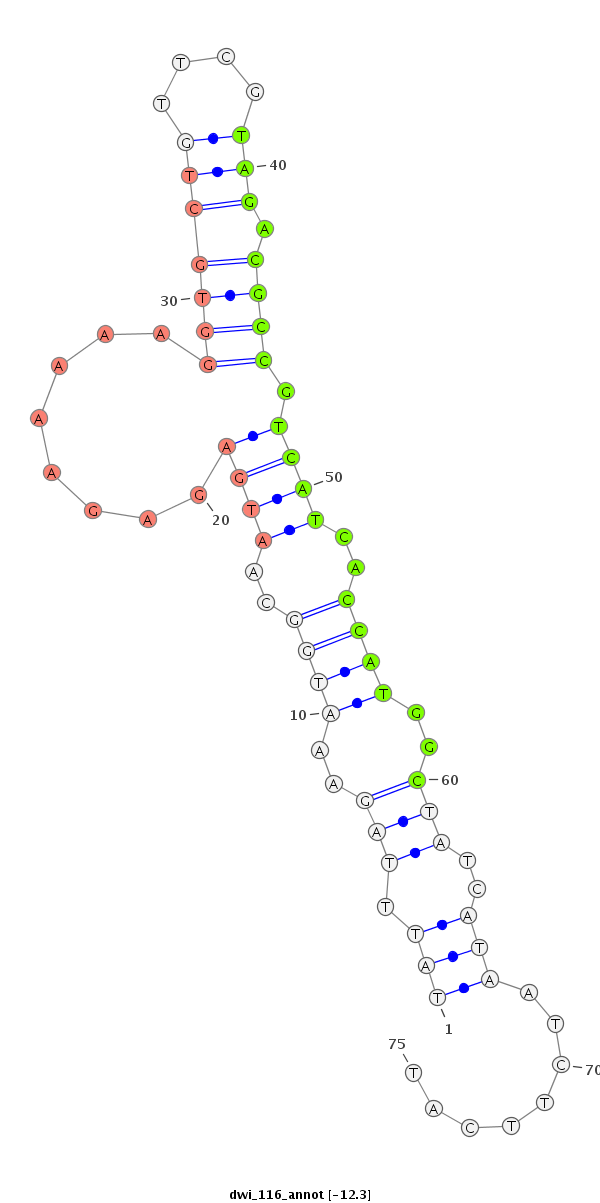
ID:dwi_116 |
Coordinate:scf2_1100000004492:119-167 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
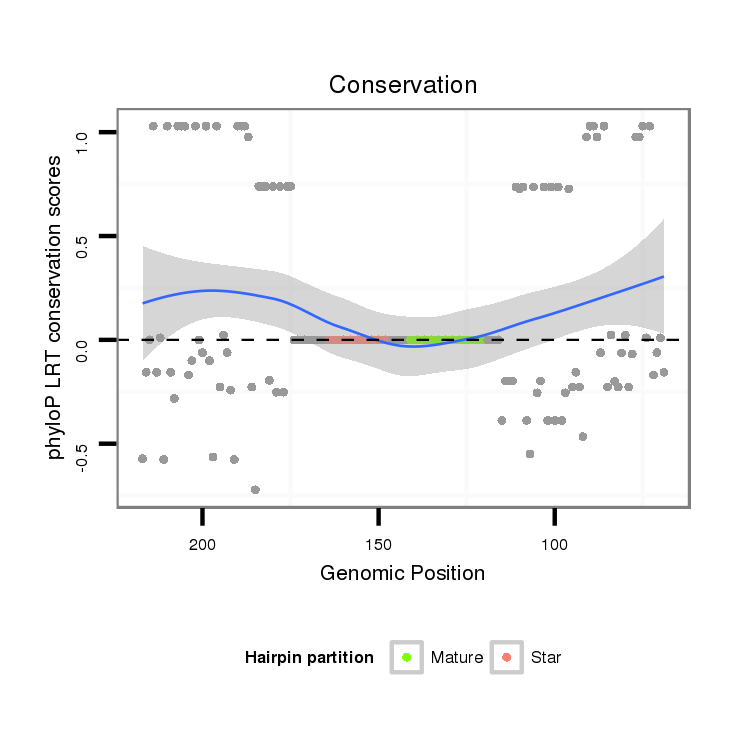
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
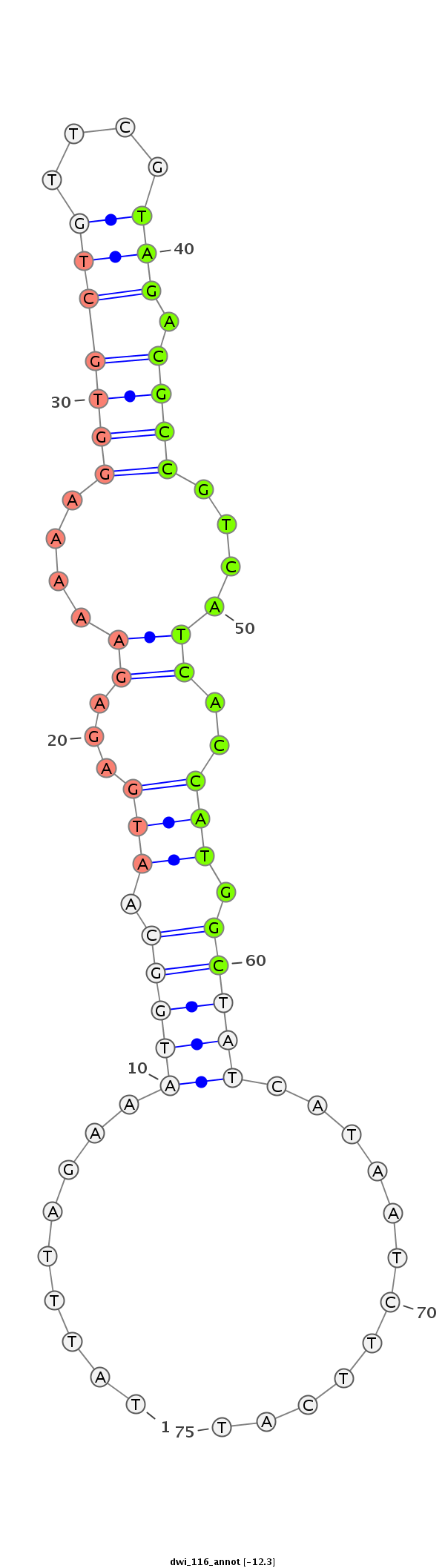
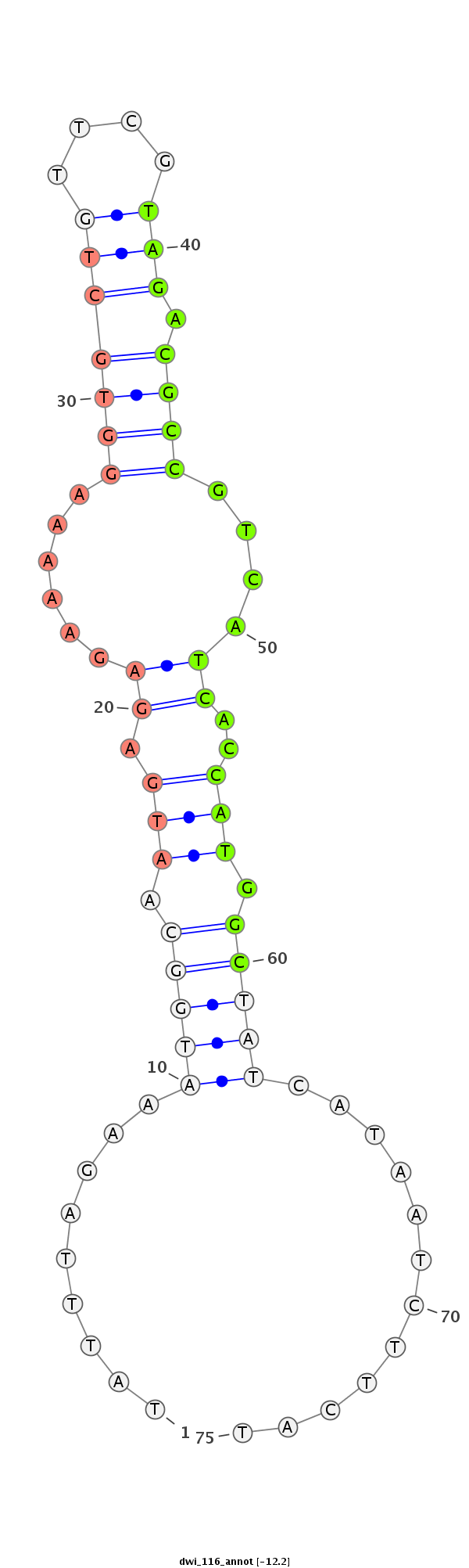
| -12.3 | -12.3 | -12.2 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CTTTTGCTACTAAACAAGAGTATAGTGATACTGTTAAATATTTAGAAATGGCAATGAGAGAAAAAGGTGCTGTTCGTAGACGCCGTCATCACCATGGCTATCATAATCTTCATCATAGTCACTTATCAAGGAAACTTTTGGCTGAAGGT
**************************************....(((..((((..((((........(((((((....))).)))).))))..))))..))).............************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
M045 female body |
M020 head |
V117 male body |
V119 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................TAGACGCCGTCATCACCATGGC................................................... | 22 | 0 | 1 | 12.00 | 12 | 11 | 1 | 0 | 0 | 0 |
| ............................................................................TAGACGCCGTCATCACCATG..................................................... | 20 | 0 | 1 | 8.00 | 8 | 6 | 2 | 0 | 0 | 0 |
| ............................................................................TAGACGCCGTCATCACCAC...................................................... | 19 | 1 | 1 | 6.00 | 6 | 3 | 2 | 0 | 1 | 0 |
| ............................................................................TAGACGCCGTCATCACCAT...................................................... | 19 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATCTTCATCATAGTCACTTA........................ | 20 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 |
| .............................................................................AGACGCCGTCATCACCATGGC................................................... | 21 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 |
| ............................................................................TAGACGCCGTCATCACCACC..................................................... | 20 | 2 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 |
| .............................................................................AGACGCCGTCATCACCAC...................................................... | 18 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................GTAGACGCCGTCATCACCAT...................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| .............................................................................AGACGCCGTCATCACCATG..................................................... | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .....................................................ATGAGAGAAAAAGGTGCTGTTC.......................................................................... | 22 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................CGTAGACGCCGTCATCACCACC..................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................ATGAGAGAAAAAGGTGCT.............................................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................TAGACGCCGTCATCACCA....................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................TAGACGCCGTCATCACAATGGC................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................GCGACGCCGTCATCACCATG..................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................GAGAAAAAGGTGCTGTTCGC........................................................................ | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................GTAGCCGCCGTCACCACCAT...................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......TACTAAACAAGAGTATAGTGA......................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................TAGACGCCGTCATCACCATGGCC.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................AAATGGCAATGAGAGAAAA..................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................TCATCATAGTCACTTATCAA.................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................AATGAGAGAAAAAGGTGCTGTTC.......................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......TACTAAACAAGAGTATAGTG.......................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........ACTAAACAAGAGTATAGT........................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GCAATGAGAGAAAAAGGTGCTGTTC.......................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................AGACGCCGTCATCACCATTGC................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................AGACGCCGTCATCACCACC..................................................... | 19 | 2 | 3 | 0.67 | 2 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................CGTAGACGCCGTCATCACCACCA.................................................... | 23 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 |
|
GAAAACGATGATTTGTTCTCATATCACTATGACAATTTATAAATCTTTACCGTTACTCTCTTTTTCCACGACAAGCATCTGCGGCAGTAGTGGTACCGATAGTATTAGAAGTAGTATCAGTGAATAGTTCCTTTGAAAACCGACTTCCA
************************************....(((..((((..((((........(((((((....))).)))).))))..))))..))).............************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
V119 head |
M045 female body |
V117 male body |
|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................CATCTGCGGCAGTAGTGGTAC..................................................... | 21 | 0 | 1 | 40.00 | 40 | 29 | 8 | 2 | 1 |
| ..........................................................................CCATCTGCGGCAGTAGTGGTAC..................................................... | 22 | 1 | 1 | 12.00 | 12 | 11 | 0 | 1 | 0 |
| ...........................................................................CATCTGCGGCAGTAGTGG........................................................ | 18 | 0 | 1 | 5.00 | 5 | 4 | 0 | 1 | 0 |
| .........................................................................AGCATCTGCGGCAGTAGTGG........................................................ | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ........................................................................................................TTAGAAGTAGTATCAGTGAA......................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ............................................................................ATCTGCGGCAGTAGTGGTAC..................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...........................................................................CATCTGCGGCCGTAGTGGTAC..................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................TGGTACCGATAGTATTAGAAG...................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................ATTAGAAGTAGTATCAGTGAA......................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ....................................................................CGACAAGCATCTGCGGCAGTAGT.......................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................TACCGATAGTATTAGAAG...................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................CGGCAGTAGTGGTACCGAT................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................AAGCATCTGCGGCAGTAG........................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................CGACAAGCATCTGCGGCAGTAG........................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................TACTCTCTTTTTCCACGACAA........................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ......................................................................................................CATTAGAAGTAGTATCAGTGAA......................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................CATCTGCGGCAGTAGAGGTAC..................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................CATCTGTGGCAGTAGTGG........................................................ | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................CCGTTACTCTCTTTTTCCACGACAA........................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................TAGAAGTAGTATCAGTGAA......................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................ACTCTCTTTTTCCACGACAA........................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................GGCAGTAGTGTTGACGATAGT.............................................. | 21 | 3 | 5 | 0.80 | 4 | 3 | 0 | 1 | 0 |
| .........................................................................ACCTTCTGCGGCAATAGTGG........................................................ | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................GTTGTTCCTTTGGAAACCGAC..... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004492:69-217 - | dwi_116 | C-----------------TTTTGCTACTAAACAAGAGTATAGTGATACTG---------------------------------------TTAAATATTTAGAAATGGCAATGAGAGAAAAAGGTGCTGTTCGTAGACGCCGTCATCACCATGGCTATCATAATCTTCATCATAGTCACTTATCAAGGAAACT--TTTG--------GCTGAAGGT------------------- |
| droAna3 | scaffold_12940:55452-55606 + | AGCAGGAGTAAAGCTTAACATCATTGATAAAAATAATAAAGATAATACACCTTTACACTATGCTGTTGAAAGGGATAAAAAAGAAATAGTTAGAAAATT-------------------------------------------------------------------------------ACTGCAAGAATGGAAGGCAGATGTTAACGCTAAAAACAATAAAGGTGATACACCTT | |
| droRho1 | scf7180000763547:9622-9767 + | TCGAGGTGC------CAATATTAATACTAAGAATGATGATGGATATACTTCTCTGCACTTTGCTGTACTGAAGAATTT--------------------------------------------------------------------------------CCGGTCTGGAATAGATTATCTTATCAAGGAAGGT--GCTAACATTGATGCTAAGGATAATATTGGTCGGGCCCCTC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/15/2015 at 02:39 PM