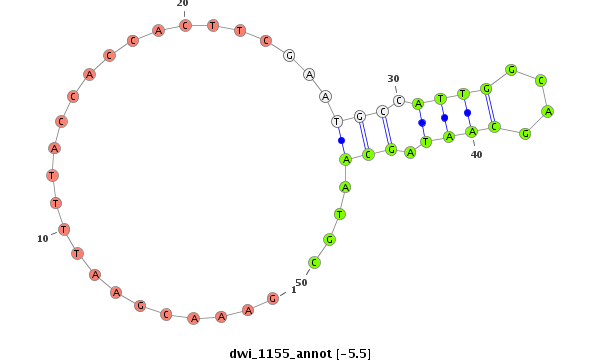
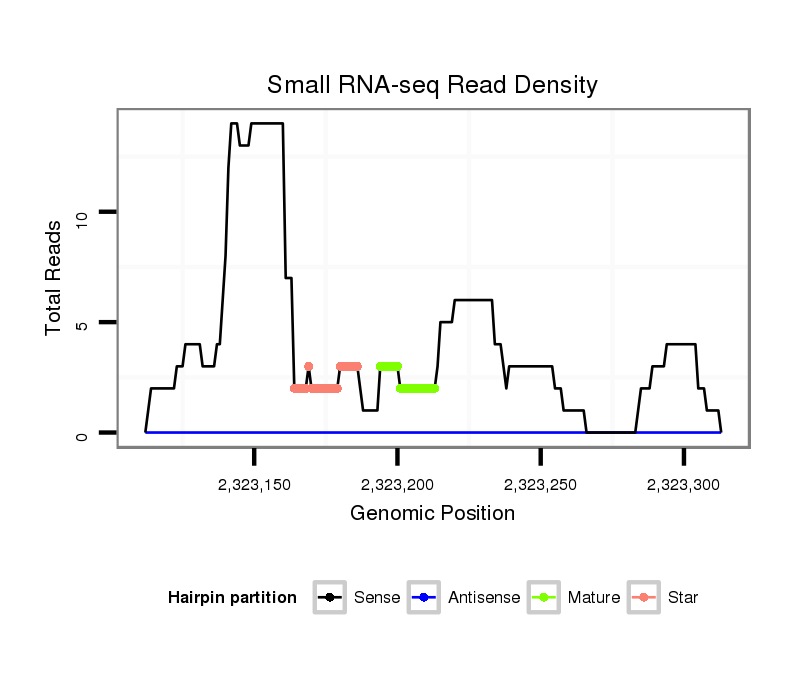
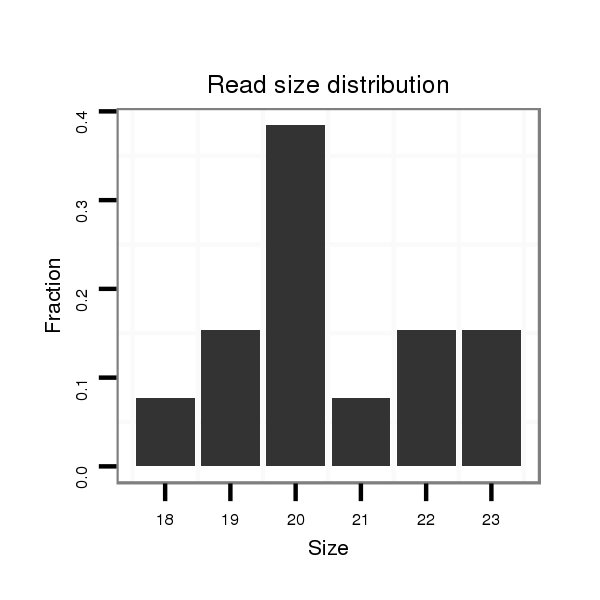
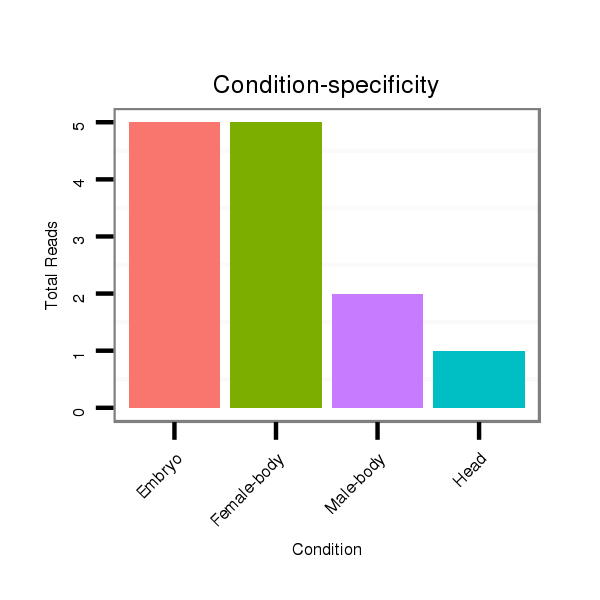
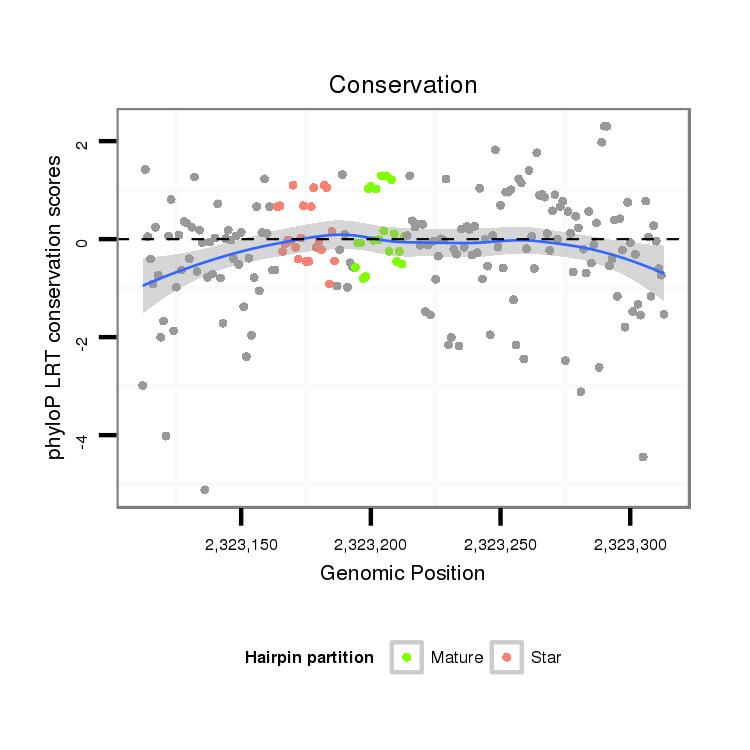
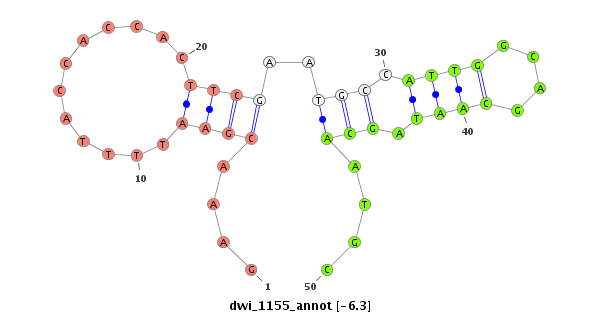
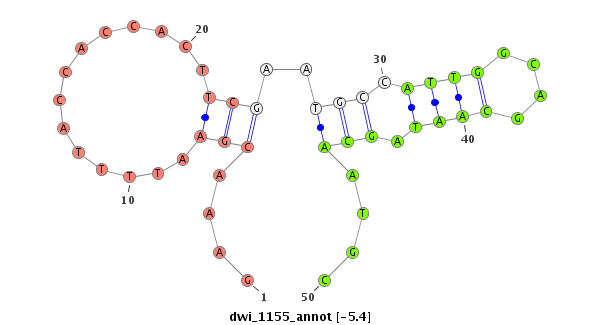
ID:dwi_1155 |
Coordinate:scf2_1100000004516:2323162-2323263 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -6.3 | -5.4 |
 |
 |
CDS [Dwil\GK23912-cds]; exon [dwil_GLEANR_7866:5]; CDS [Dwil\GK23912-cds]; exon [dwil_GLEANR_7866:6]; intron [Dwil\GK23912-in]
No Repeatable elements found
| ##################################################------------------------------------------------------------------------------------------------------################################################## CAAGGAACGCAAAGAGAAGAAACGTGAGAAATTGAGGAATTTGGCTGGCGGCGAAACGAATTTTACCACCACTTCGAATGCCATTGGCAGCAATAGCAATGCCATTTGTGGTGTTCCCAGTGGTGCTACTAATGCCACCACCAGCACGACAACAACAGCAACATCAACAACCACTACCAACGATATCAGCAGCGAGAAACTG ****************************************************..........................(((.((((....)))).)))....**************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
V118 embryo |
M020 head |
V119 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ................................TGGGGAGTTTGGCTGGCGG....................................................................................................................................................... | 19 | 2 | 3 | 31.33 | 94 | 72 | 22 | 0 | 0 | 0 |
| ...............................GTGGGGAGTTTGGCTGGCGG....................................................................................................................................................... | 20 | 3 | 11 | 18.55 | 204 | 174 | 29 | 1 | 0 | 0 |
| ...............................GTGGGGAGTTTGGCTGGCGGC...................................................................................................................................................... | 21 | 3 | 4 | 16.75 | 67 | 35 | 31 | 0 | 0 | 1 |
| ................................TGGGGAGTTTGGCTGGCGGC...................................................................................................................................................... | 20 | 2 | 3 | 11.33 | 34 | 18 | 14 | 2 | 0 | 0 |
| ................................TGGGGAGTTTGGCTGGCGGCA..................................................................................................................................................... | 21 | 3 | 6 | 5.17 | 31 | 27 | 4 | 0 | 0 | 0 |
| .................................GGGGAGTTTGGCTGGCGG....................................................................................................................................................... | 18 | 2 | 8 | 2.63 | 21 | 15 | 6 | 0 | 0 | 0 |
| .............................AATTGAGGAATTTGGCTGGCGGC...................................................................................................................................................... | 23 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 |
| ..................................................................................ATTGGCAGCAATAGCAATGC.................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 |
| ..............................ATTGAGGAATTTGGCTGGC......................................................................................................................................................... | 19 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...........................GAAATTGAGGAATTTGGCTGGCGGC...................................................................................................................................................... | 25 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| .............................AATTGAGGAATTTGGCTGGC......................................................................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ................................TGGGGAGTTTGGCGGGCGGC...................................................................................................................................................... | 20 | 3 | 7 | 1.29 | 9 | 4 | 5 | 0 | 0 | 0 |
| ......................................................................................................................................................................................ATATCAGCAGCGAGAAACT. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................AAATTGAGGAATTTGGCTGGC......................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................GAAACGAATTTTACCACCACTTC............................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................GAAATTGAGGAATTTGGCTGGC......................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................CCACTTCGAATGCCATTGGCA................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................ACTAATGCCACCACCAGCACGACAACA................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........AAGAGAAGAAACGTGAGAAATT......................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................GAATTTTACCACCACTTCG.............................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GCTACTAATGCCACCACCAGCA........................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................GGGGAGTTTGGCTGGCGGC...................................................................................................................................................... | 19 | 2 | 5 | 1.00 | 5 | 4 | 1 | 0 | 0 | 0 |
| ............................................................................................................TGGTGTTCCCAGTGGTGC............................................................................ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................ACTACCAACGATATCAGCAGC......... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................AAATTGAGGAATTTGGCTGGCGGC...................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..AGGAACGCAAAGAGAAGAAACGTGA............................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................GAGAAATTGAGGAATTTGGCTGGC......................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............AGAAGAAACGTGAGAAATT......................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................CATTTGTGGTGTTCCCAGTGGTG............................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................ATTTGTGGTGTTCCCAGTG................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................GAGGAATTTGGCTGGCGGC...................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................ATTTGTGGTGTTCCCAGTGG............................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................CTACCAACGATATCAGCAGC......... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................AATTTGGCTGGCGGCGAAACG................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................CATTTGTGGTGTTCCCAGTGGT.............................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .AAGGAACGCAAAGAGAAGA...................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................CAACGATATCAGCAGCGAG...... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................GGGAGTTTGGCTGGCGGC...................................................................................................................................................... | 18 | 2 | 8 | 1.00 | 8 | 2 | 6 | 0 | 0 | 0 |
| ...........................................................................................................................TGCTACTAATGCCACCACCA........................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................CATTTGTGGTGTTCCCAGTG................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................TGGGGAGTTTGGCTGGGGGC...................................................................................................................................................... | 20 | 3 | 13 | 0.62 | 8 | 8 | 0 | 0 | 0 | 0 |
| .............................AGTTGGGGAGTTTGGCTGGCGG....................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................AGTGGGGAGTTTGGCTGGCGGC...................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................TTGGGGAGTTTGGCTGGCGG....................................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................AGTTGGGGAGTTTGGCTGGCG........................................................................................................................................................ | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................GAATTTGGGTAGGGGCGAAA.................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................GTTGGGGAGTTTGGCTGGCGG....................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................GGGGAGTTTGGCTGGGGGCG..................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................GTGGGGATTTTGGCTGGCGG....................................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................ATGCCATTGGCCGCAGTAGA......................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................TTGGGAGTTTGGCTGGCGGC...................................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GTGTTTCCAGTGGTGCAATT........................................................................ | 20 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................GAATTTGGGTTGGGGCGAAA.................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................GGTGAGGAGTTTGGCTGGC......................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................TGGGGAGTTTGTCTGGCGG....................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
GTTCCTTGCGTTTCTCTTCTTTGCACTCTTTAACTCCTTAAACCGACCGCCGCTTTGCTTAAAATGGTGGTGAAGCTTACGGTAACCGTCGTTATCGTTACGGTAAACACCACAAGGGTCACCACGATGATTACGGTGGTGGTCGTGCTGTTGTTGTCGTTGTAGTTGTTGGTGATGGTTGCTATAGTCGTCGCTCTTTGAC
****************************************************************************************************..........................(((.((((....)))).)))....**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
M020 head |
V117 male body |
|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................TTACGATAGGGGTCGTGCTG.................................................... | 20 | 3 | 4 | 0.75 | 3 | 3 | 0 | 0 |
| ........................................................................................................................................................................................TAGTCGTGGATCTTTGAC | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ...........................................................TTAAATGGTGGTGAATCT............................................................................................................................. | 18 | 2 | 15 | 0.13 | 2 | 1 | 0 | 1 |
| ....................................................................................................................................ACGGGGGTGGGCGTGCTG.................................................... | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 |
| ......................................................................................................................................GGTGGGGGTGGTGCTGTTGTT............................................... | 21 | 2 | 11 | 0.09 | 1 | 0 | 0 | 1 |
| ..........................................................................................................ACACGACACGGGTCGCCAC............................................................................. | 19 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................................TGTTGTTGTCGTTGTAGTT................................... | 19 | 0 | 18 | 0.06 | 1 | 0 | 0 | 1 |
| ...................................................................................................................................TAAGGTGGTAGTCGTGCGG.................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004516:2323112-2323313 + | dwi_1155 | CAAGGAACGCAAAGAGAAGAAACGTGAGAAATTGAGGAATTTGGCT-------------------------------------------------------------------------------------GGCGGCGAAACGAATTTTACCACCACTTCGAATGCCATT----------------------GGCAGCAATAGCAATGCCA--------------------------------------------------TTT-----------------------------------------------------------------------GTG------GTGTTCCCAGTGGTGCTACTAAT-------G------CCA------CCACCAGCACG-------------------------------------------------------------ACAACAA------C---AGCAACATCAACA----------------------------------ACCACT---ACCAAC----G---ATATCAG----------CAGCGAGAAACTG |
| droVir3 | scaffold_12875:15671131-15671342 + | TAAAAAACAGAACGAAAAAAATTTGAGTAAGCAGTT-GGCTGTC-GGCAGTTTATGACCTTTTCGAAAGGTGGCGACAGGCTGCTGATTGCAGCTTAGCCTTAATAATGGCCCACAAAACGGTTTTAGACCAGC---------------------------T---------------AGTAAAGCTTT---AAA----------------------------TAA-----------------------CCT---TAAA-----------------------------------------------------------------------------------------------------------------GTA----ACAGCAACAGCAAC------------------------------------------------------------------------AGCAA------C---AACAACATC------------------------------------------------------------------AACAAACATCAGAGTCGAATAGC-- | |
| droMoj3 | scaffold_6473:14568994-14569171 + | GAAAAGGGGAAAAAAGAGCAACTGCT-GAGGACGAGGCGAACAA-----------------------------------------------------------------------------------------------------------------CAACTATGAATAAGGCAGGCGGA-------------------ACAGCCGCACGA--------------------------------------------------TTCG----------------------CTA--------------------AAT----------------------CCGAATTGAAATGT--------TGCCGCC---TCG----GCTAGCAGCAACAGCAAC------------------------------------------------------------------------AACAA------C---AACAACATGTTAA----------------------------------ACTAGC---AACAAC----A---ACAACAG--------------CAACTATAA | |
| droGri2 | scaffold_14846:183762-183975 - | CAAAGGAGGCAATAAAAA---ATTCAAAAACTTGAA---TCTGGCC-------------------------------------------------------------------------------------AGAGCGGAGGTGATGTTGCCCTCAACGTCGAAACTCA------------------------AGCAACAACAACAATTGCAACTTAATTGCCC--------------------------------------TTC-----------------------ATC--------------------AT---------------------------------------CCTC-------------------------------------------------------------------ATCGTCAACTGCTGCTGCTGC---TGTTGCTGCTCCTACGACAACGA------CAAC---AACAGCAGCA---A------CTATAGCGAGTG--CGAGTGC--AA-CGCT----------------------GG--------------CAGCGA-CG | |
| dp5 | 4_group1:4426724-4426940 - | dps_1419 | CAAGGAGCGCAAGGAGAAGAAACGCGAGAAGATGAGGAACCTCA--------------------------------------------------------------------------------------------------------------------------------------------CGGCGGAGAC----------------------------AAACACCATCGCTG-----------------------------GGTCTGCCACCA-----------------------------------------------------------------------------------------------------------------------------CAGCAGCAGCACCATCACACATGAGCAGCGGCAGCAGCAGC-ACACCAGCTGCACCGCCAGCAGCAA------CAGC---AGCAGCAGCG---GCTGT------G---------CCGGTTC-CGTCTGCCGGCAGCAACGATCTCGGGGAAAAG-----------CTGGAGAAGCTA |
| droPer2 | scaffold_5:783361-783577 + | CAAGGAGCGCAAGGAGAAGAAACGCGAGAAGATGAGGAACCTCA--------------------------------------------------------------------------------------------------------------------------------------------CGGCGGAGAC----------------------------AAACACCATCGCTG-----------------------------GGTCTGCCACCA-----------------------------------------------------------------------------------------------------------------------------CAGCAGCAGCACCACCACACATGAGCAGCGGCAGCAGCAGC-ACACCAGCTGCACCGCCAGCAGCAA------CAGC---AGCAGCAGCG---GCTGT------G---------CCGGTTC-CGTCTGCCGGCAGCAACGATCTCGGGGAGAAG-----------CTGGAGAAGCTA | |
| droAna3 | scaffold_12943:4205269-4205395 - | CAAGGAGCGGAAGGAGAAGAAGCGAGAGAAGCTGAGGAACCTCA--------------------------------------------------------------------------------------------------------------------------------------------CC---GAGGC----------------------------CAA-----------------------CAG---CACCACT----------------------------------------------------------------------------------------------------------GCGAACA----GCAGCAACTCCACCA-------------------------------------------------------------------------------------------------------------CCAA---------------CTC-CGCCACC---ACCAAT----G---ATGTGAC----------TTCAGAGAAACTC | |
| droBip1 | scf7180000395973:424936-425065 - | CAAGGAGCGCAAGGAGAAGAAGCGCGAGAAGCTGAGGAATCTCA--------------------------------------------------------------------------------------------------------------------------------------------CC---GAGGC----------------------------CAA-----------------------CAGTGCCACCACA----------------------------------------------------------------------------------------------------------GCGATTG----GCGGCAACTCCACCG-------------------------------------------------------------------------------------------------------------CCAA---------------CTC-CGCCACA---CCCAAC----G---ATGTGAC----------ATCCGAGAAGCTC | |
| droKik1 | scf7180000302273:156753-156888 + | TAAGGAGCGCAAGGAGAAGAAGCGCGAGAAGCTGAGGAACCTCA--------------------------------------------------------------------------------------------------------------------------------------------CA---GAGAC----------------------------CAACGCAACCGTTG-----------------------------GAAGCG-----------------------------------------------------------------------------------------------------------------------------TGACCACAGTA--A-----------------------------------------------------------------A---CACAA------------------CCAA------TCCTCC---AAT-GGTTACA------AAT----GAGGTGATTAG----------CTCTGAGAAACTT | |
| droFic1 | scf7180000453316:88080-88249 + | GAG----TTCAATGCAAAGACATGTGCATAAATGAAGAGCTACA--------------------------------------------------------------------------------------------------------------------------------------------GC---CAATT----------------------------TAATTTAAACGCCG---------CCG-------------CCC-CGCCCGCCATTG--------CATACGACAATTCAGTTGTAAATCCAAACGG-------------------TTCGCCTTTAGA----------------------------------T------------------------------------------CAGCAACAACAAC------------------------------AA------C---AGC---------A---A------CAACAACAAGCG--------------CAGC----------------------AG--------------CAGCAG-CG | |
| droEle1 | scf7180000490216:115762-115879 + | CAAGGAGCGGAAGGAAAAGAAACGCGAGAAACTGAGGAATATGG--------------------------------------------------------------------------------------------------------------------------------------------CA---GATAC----------------------------CAA-----------------------TAACAGCACCACC----------------------------------------------------------------------------------------------------------GTAAG-------C---------GCCA-------------------------------------------------------------------------------------------------------------CTAA---------------TAC-TCCTACC---ACCAAT----G---ATATTAG----------CTTGGAGAAATTG | |
| droRho1 | scf7180000777787:21642-21805 - | AACAGACAACAAGCGAAAGCAACAACAACAACA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGAGGGCGTTGGCT-------------C-----------ACCAGGCAACCACACAC--------------------AATCACAC----------------------------------------------------------------------------------------------------------CAACAACAGCAGCAACAGCAACAACAGCTGCATCA-------CAACAACAACACC---AGCAACAACAGCAAT--T------G---------CCACCAC--AGTCACC----------------------AG--------------CAGCAACAA | |
| droBia1 | scf7180000301665:2004-2129 - | CCACCAACATCGGCAGCC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCAGCTACAACATCGCCAGCTGAAACGCCC---------------------------------------------------------------------------------------------------------------------------------AAAGGCGCCGCTAA-------------CATCA------------------------------------------------------------------------ACGCCAGCGCCAA------C---AACAACATCGGCA----------------------------------GCCAGC---GCCAAC----A---ACATCGG----------CAGCCAGCAAC-- | |
| droTak1 | scf7180000414118:36918-37089 - | AC------CCAAAAAAACGAAACGAAAGAAAATAAGAGAGGCGG--------------------------------------------------------------------------------------------------------------------------------------------CTACGAATTC----------------------------GAA----------------------------------------------------------TATGTTC--------------------AATCATACCAAACTTTTTTCAGATTTTTGATTTTGTATCCCCATCATCACCACCACC-------A------CTA------CCACCACAACT-------------------------------ATAAC------------------------------AA------C---AAC---------A---A------CCAG---------------TAC-AAGCACC---AACAAC----A---ACATCAA----------C------------ | |
| droEug1 | scf7180000409154:180218-180329 + | CAAGGAAAGAAAAGAAAAGAAACGTGAGAAGTTGCGGAATATAA--------------------------------------------------------------------------------------------------------------------------------------------CG---GAGAT----------------------------CAA-----------------------TAGCATCACTAC--------------------------------------------------------------------------------------------------------------------------------TATAG-------------------------------------------------------------------------------------------------------------GCAA---------------TTC-GCCTACC---AGCAAT----G---ATTTAAG----------TTCTGAAAAATTA | |
| dm3 | chr2L:11255602-11255739 + | GATGCAGCAGCAGGAGCAGCAGCAGCGTCAACTGTTTGACC------------------------------------------------------------------------------------------AGC-------------------------------------------GGCGCAGCCAC---CAC----------------------------CAG-----------------------CCT---CACCATC----------------------------------------------------------------------------------------------------------ACCACCA----GCCGCATCATCATCG---CCACCACC------------------------------------------------------------------------------ATCAT----C---A--CC-AGCAA------CG--------------CATC---AACAAC-------------AA--------------TAACAATAA | |
| droSim2 | 3r:15722632-15722820 + | AATGAAAGA-AAAAAAAAAAAACGAAAGAAAATAAGAGACGCCA----------------------------------------------------------------------------------------G---------------------------------------------------CTACGAATTC----------------------------GAA----------------------------------------------------------TATGTTC--------------------AATCATACCAAACTTTTTTCAGATTTTTGATTTTGTATCCCCATCATCACCACCACC-------A------CTA------CCACCACAACT-------------------------------------------------------------------ACAACAAC---AACAAC------A---A------CCAG---------------TAC-AAGCACC---AACAAC----A---ACATCAA--------------CAACAACAA | |
| droSec2 | scaffold_3:6259721-6259888 - | AATTTGAATTT---GAACCAAAAGCGTTTGGCCGGTGCT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCT-----------------------------A--------------------------------------------------------------------------------ATCCCCTGCAGCAACAA---------------------------------------------CAACAGCATC-CACAGCAACAGCTTCAACAAC------AACAACAACAACAACAACAACAA------CAAC---AACAACAACA---ACAAC------A---------TCAACAT-CATC-AACAACAGCAACAG-----------------------G-TCATGCAATTG | |
| droYak3 | 2R:17620353-17620491 + | AATTAAAATATTTTGGCATCAG---------------------T------------------------------------------------------------------------------------------------ACTAATC----CAACACTTTCTATTCACTTTGCAGTTGG-------------------------------------------------------------CGCTGGCG-------------CTC-----------------------------------------------------------------------TTGGCCTTTGGAGTTGCCTCATC-----------A----TC---------CTCCACCACCAGCACA-------------------------------------------------------------ACCACAT------C---GGTGAAGCCAA--------------------------------------------------------------------------------AGACCAATA | |
| droEre2 | scaffold_4929:237579-237678 + | CAAGGAAAGGAAGGAGAAGAAACGTGAGAAGCTTAGGAACATGA--------------------------------------------------------------------------------------------------------------------------------------------CG---GAATC----------------------------CA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGTACCCAA---------------TTC-GCCTACC---ACCAAT----G---ACACAAG----------TTCCGAAAAATTA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 12:46 AM