| GGAAATC---A----------AAT---TCAAAGG------------------------------AGCC----------TC-TACCAGCAGCAGA------AGC---------AG----------C---------AACAACAGCAACAGCAGTA---------GCAACAGC---A---------------------GCAAC---AACAACAGCAGCAGCAA------------------------------CAACGA--------CG------ACAT------CAG---------CTA-------------------------------AGTCAAAACAAGAATTTTC----------------------TGTTTATAC---------------------AAA--CACACATAT--ATAACAA------------------------------A-------------AA---------------------------------------AAAAATATACAGTCCA | Size | Hit Count | Total Norm | Total | M055
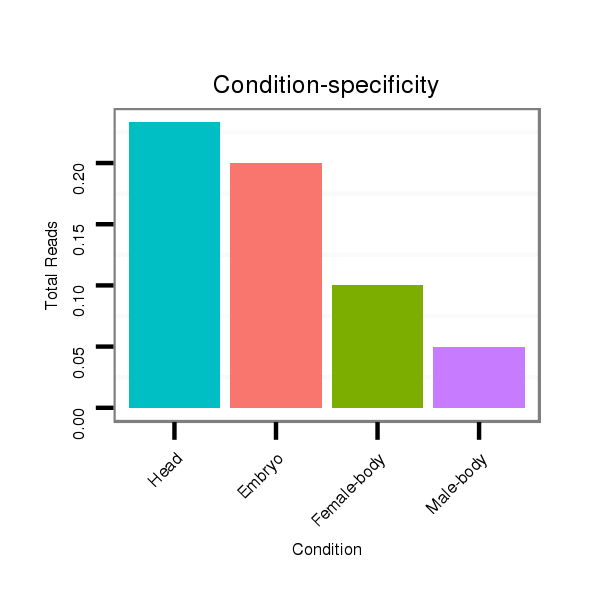
Female-body | M057
Embryo | V060
Head | V040
Embryo | V107
Male-body | V108
Head | GSM1528801
follicle cells |
| ....ATC---A----------AAT---TCAAAGG------------------------------AGCC----------TC-TACC...................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 6 | 1.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ATC---A----------AAT---TCAAAGG------------------------------AGCC----------TC-TACCAGC................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 6 | 0.50 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................AACGA--------CG------ACAT------CAG---------CTA-------------------------------AGTCAAAT........................................................................................................................................................................................... | 25 | 6 | 0.50 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| ....ATC---A----------AAT---TCAAAGG------------------------------AGCC----------TC-TACCAGCA.................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 28 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................AGTCAAAACAAGAATTTTC----------------------TGTTT..................................................................................................................................................... | 24 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ATC---A----------AAT---TCAAAGG------------------------------AGCC----------TC-TACCA..................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TA---------GCAACAGC---A---------------------GCAAC---AACAACAGCA...................................................................................................................................................................................................................................................................................................................... | 26 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AAC---AACAACAGCAGCAGCAA------------------------------CAACGA--------CG------........................................................................................................................................................................................................................................................... | 28 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................GCAGTA---------GCAACAGC---A---------------------GCAAC---AACAAC.......................................................................................................................................................................................................................................................................................................................... | 26 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TCAAAGG------------------------------AGCC----------TC-TACCAGCAGCAGA------A...................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................AACAGCAGCAGCAA------------------------------CAACGA--------CG------ACA........................................................................................................................................................................................................................................................ | 25 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................AAGG------------------------------AGCC----------TC-TACCAGCAGCAGA------....................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................T---TCAAAGG------------------------------AGCC----------TC-TACCAGCAGCAGA------AGC---------........................................................................................................................................................................................................................................................................................................................................................................................................................... | 30 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................TC-TACCAGCAGCAGA------AGC---------AG----------C---------AACAA................................................................................................................................................................................................................................................................................................................................................................................................ | 26 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................AAGG------------------------------AGCC----------TC-TACCAGCAGCAGA------AGC---------A.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TA---------GCAACAGC---A---------------------GCAAC---AACAAC.......................................................................................................................................................................................................................................................................................................................... | 22 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................AAAGG------------------------------AGCC----------TC-TACCAGCAGCAGA------AGC---------........................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................C----------TC-TACCAGCAGCAGA------AGC---------AG----------C---------AACAACA.............................................................................................................................................................................................................................................................................................................................................................................................. | 29 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TACCAGCAGCAGA------AGC---------AG----------C---------AACAACAGC............................................................................................................................................................................................................................................................................................................................................................................................ | 28 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AAC---AACAACAGCAGCAGC................................................................................................................................................................................................................................................................................................................. | 18 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| ..............................AAGG------------------------------AGCC----------TC-TACCAGCAGCAGA------AGC---------AG----------C---------..................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................TC-TACCAGCAGCAGA------AGC---------AG----------C---------AACA................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................T---TCAAAGG------------------------------AGCC----------TC-TACCAGCAG................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................AGCAA------------------------------CAACGA--------CG------ACAT------CAG---------CTA-------------------------------AG................................................................................................................................................................................................. | 25 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....TC---A----------AAT---TCAAAGG------------------------------AGCC----------TC-T......................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................AG---------CTA-------------------------------AGTCAAAACAAGAATTTTC----------------------.......................................................................................................................................................... | 24 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TA-------------------------------AGTCAAAACAAGAATTTTC----------------------TGTTTA.................................................................................................................................................... | 27 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................AT---TCAAAGG------------------------------AGCC----------TC-TACCAGCAAT................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................ACAGCAGCAGCAA------------------------------CAACGA--------CG------ACAT------C................................................................................................................................................................................................................................................ | 26 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................A-------------------------------AGTCAAAACAAGAATTTTC----------------------.......................................................................................................................................................... | 20 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................CTA-------------------------------AGTCAAAACAAGAATTTTC----------------------TGT....................................................................................................................................................... | 25 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....TC---A----------AAT---TCAAAGG------------------------------AGCC----------TC-TACCAG.................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................AT---TCAAAGG------------------------------AGCC----------TC-TACCAGCA.................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TA-------------------------------AGTCAAAACAAGAATTT.................................................................................................................................................................................. | 19 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................T------CAG---------CTA-------------------------------AGTCAAAACAAGAATTTTC----------------------.......................................................................................................................................................... | 26 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................AGTCAAAACAAGAATTTTC----------------------TGC....................................................................................................................................................... | 22 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....TC---A----------AAT---TCAAAGG------------------------------AGCC----------TC-TACCAGCA.................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................CG------ACAT------CAG---------CTA-------------------------------AGTCAAAACAAGA...................................................................................................................................................................................... | 25 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TA-------------------------------AGTCAAAACAAGAATTTTC----------------------TGTT...................................................................................................................................................... | 25 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....ATC---A----------AAT---TCAAAGG------------------------------AGCC----------TC-TACCAA.................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......C---A----------AAT---TCAAAGG------------------------------AGCC----------TC-TACC...................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................AG---------CTA-------------------------------AGTCAAAACAAGAATTTTC----------------------TGT....................................................................................................................................................... | 27 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ATC---A----------AAT---TCAAAGG------------------------------AGCC----------TC-TACCAG.................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................A---------GCAACAGC---A---------------------GCAAC---AACAACAGC....................................................................................................................................................................................................................................................................................................................... | 24 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................CAGCAGCAGA------AGC---------AG----------C---------AACAACAG............................................................................................................................................................................................................................................................................................................................................................................................. | 24 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................A---------------------GCAAC---AACAACAGCAGCAGC................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .....................................................................................AGCAGCAGA------AGC---------AG----------C---------AACAACAGT............................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................A---------GCAACAGC---A---------------------GCAAC---AACAACA......................................................................................................................................................................................................................................................................................................................... | 22 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................C---A---------------------GCAAC---AACAACAGCAGCAGC................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................CAAC---AACAACAGCAGCAGC................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................CAGC---A---------------------GCAAC---AACAACAGCAGCA................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................CAAC---AACAACAGCAGCAGCA................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................GC---A---------------------GCAAC---AACAACAGCAGCAGC................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................CAACAGCAGCAGCAA------------------------------CAAC............................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |