
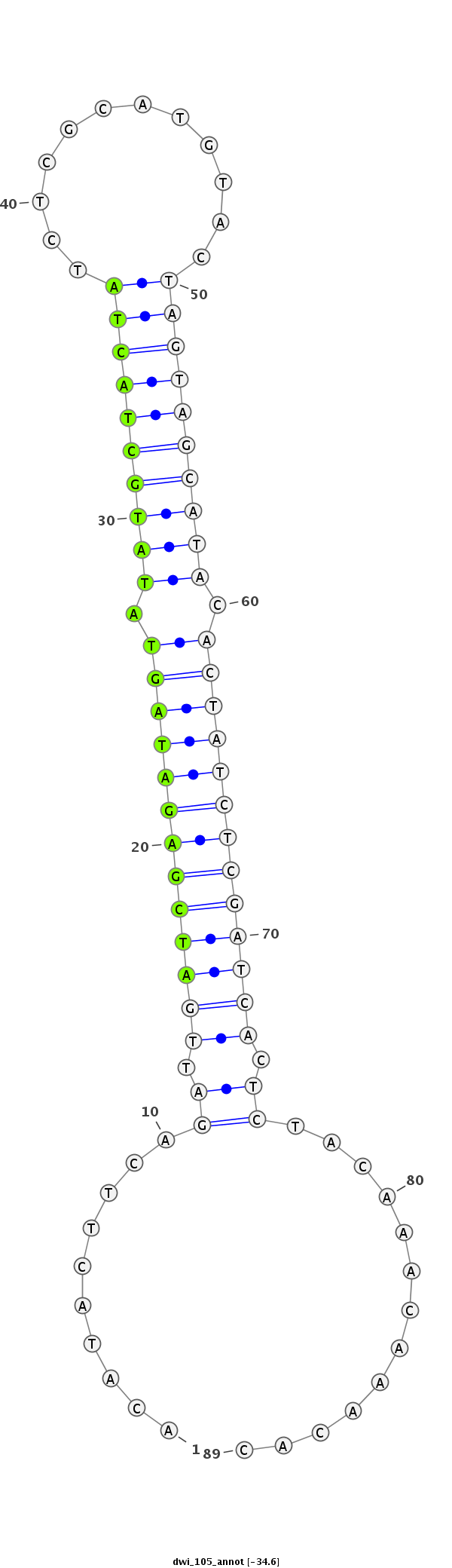
ID:dwi_105 |
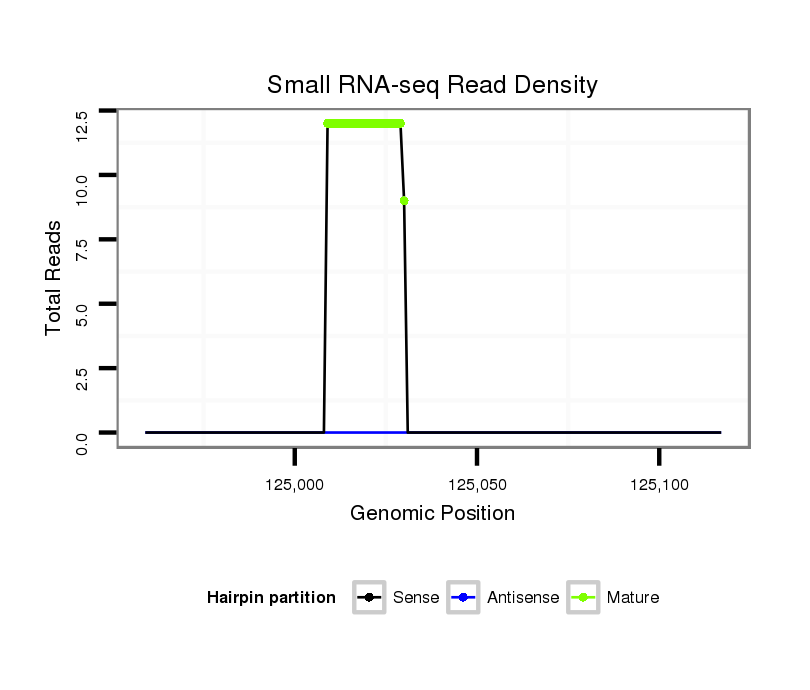
Coordinate:scf2_1100000004595:125009-125067 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -34.6 |
 |
Antisense to intron [Dwil\GK20295-in]
No Repeatable elements found
| mature | star |
|
TCGAAAACCAAATGTATGTTTACTAAATTGTTTTTACATACTTCAGATTGATCGAGATAGTATATGCTACTATCTCGCATGTACTAGTAGCATACACTATCTCGATCACTCTACAAACAAACACAGCTACTACAAACGCAGCTACTAACCAACGGACAA
***********************************.........(((.(((((((((((((.((((((((((............)))))))))).))))))))))))).)))............*********************************** |
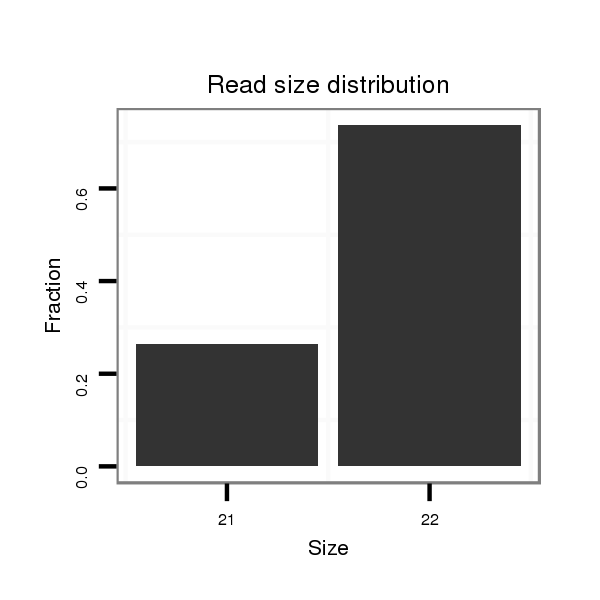
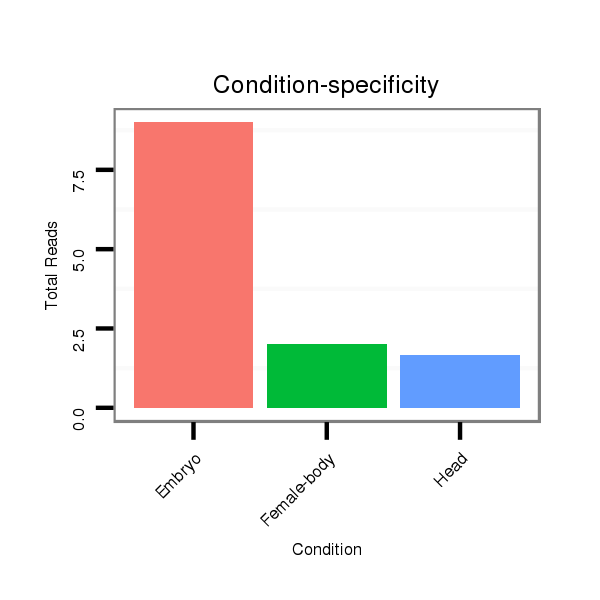
Read size | # Mismatch | Hit Count | Total Norm | Total | V118 embryo |
M045 female body |
V119 head |
|---|---|---|---|---|---|---|---|---|
| ..................................................ATCGAGATAGTATATGCTACTA....................................................................................... | 22 | 0 | 3 | 9.33 | 28 | 21 | 4 | 3 |
| ..................................................ATCGAGATAGTATATGCTACT........................................................................................ | 21 | 0 | 3 | 3.33 | 10 | 6 | 2 | 2 |
| ..................................................ATCGAGATAGTATATGCTACTAA...................................................................................... | 23 | 1 | 3 | 3.00 | 9 | 9 | 0 | 0 |
| ..................................................ATCGAGATAGTATATGATACT........................................................................................ | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ..................................................ATCGAGATAGCATATGCTACTAA...................................................................................... | 23 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ........................................................................................................................................CGCAGCTACTAACCAACGGAC.. | 21 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ......................................................................................GTAGAATACACTATCTCGATCA................................................... | 22 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ..................................................ACCGAGATAGTATATGCTACTA....................................................................................... | 22 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 |
| .CGAAAACCAAATGTATGTTTA......................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
|
AGCTTTTGGTTTACATACAAATGATTTAACAAAAATGTATGAAGTCTAACTAGCTCTATCATATACGATGATAGAGCGTACATGATCATCGTATGTGATAGAGCTAGTGAGATGTTTGTTTGTGTCGATGATGTTTGCGTCGATGATTGGTTGCCTGTT
***********************************.........(((.(((((((((((((.((((((((((............)))))))))).))))))))))))).)))............*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M020 head |
M045 female body |
|---|---|---|---|---|---|---|---|
| ............................................................................................CCGTGATAGAGCTAGTGAGATG............................................. | 22 | 2 | 3 | 0.67 | 2 | 2 | 0 |
| .........................................................................................CGTATGTGATAGAGCTAGTG.................................................. | 20 | 0 | 3 | 0.33 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004595:124959-125117 + | dwi_105 | TCGAAAACCAAATGTATGTTTACTAAATTGTTTTTACATACTTCAGATTGATCGAGATAGTATATGCTACTATCTCGCATGTACTAGTAGCATACACTATCTCGATCACTCTACAAACAAACACAGCTACTACAAACGCAGCTACTAACCAACGGACAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
Generated: 05/15/2015 at 02:40 PM