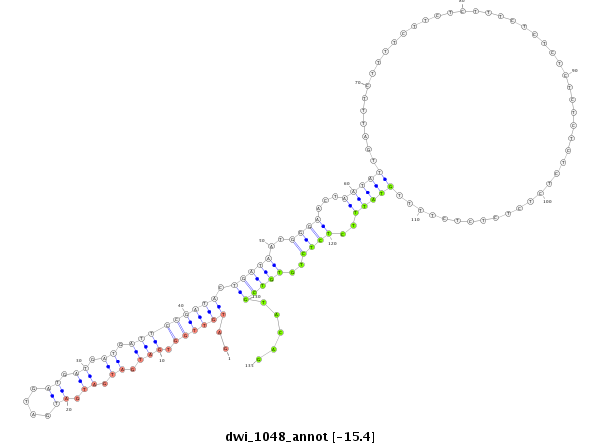
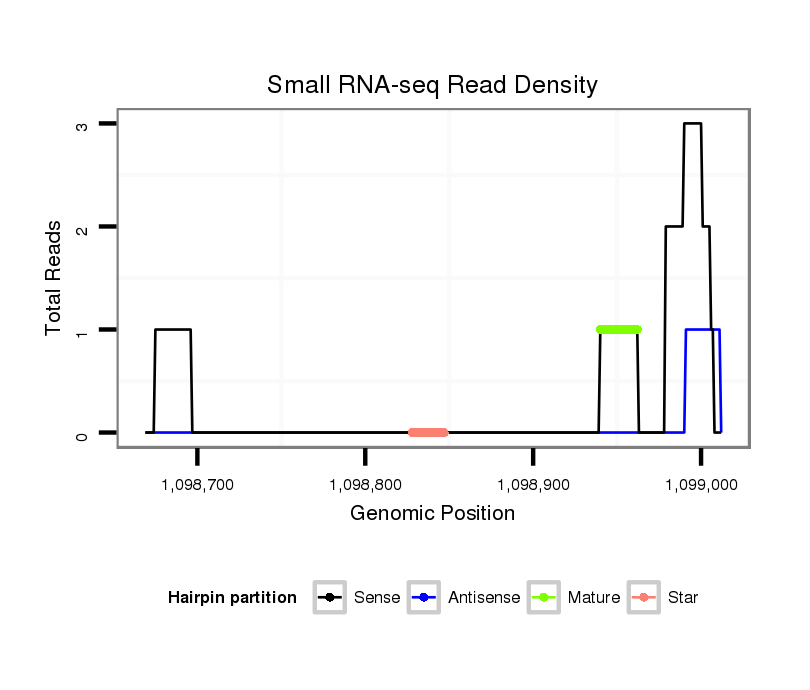
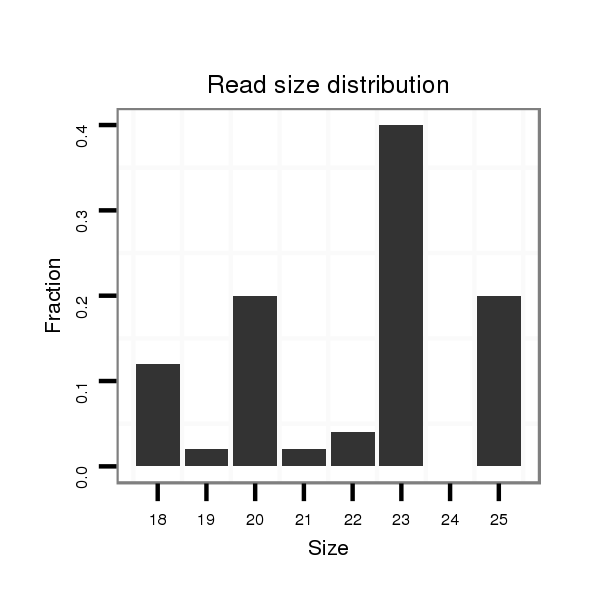
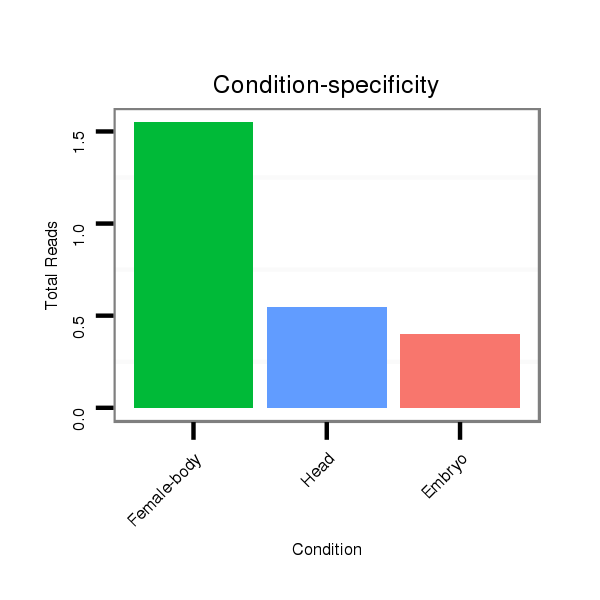
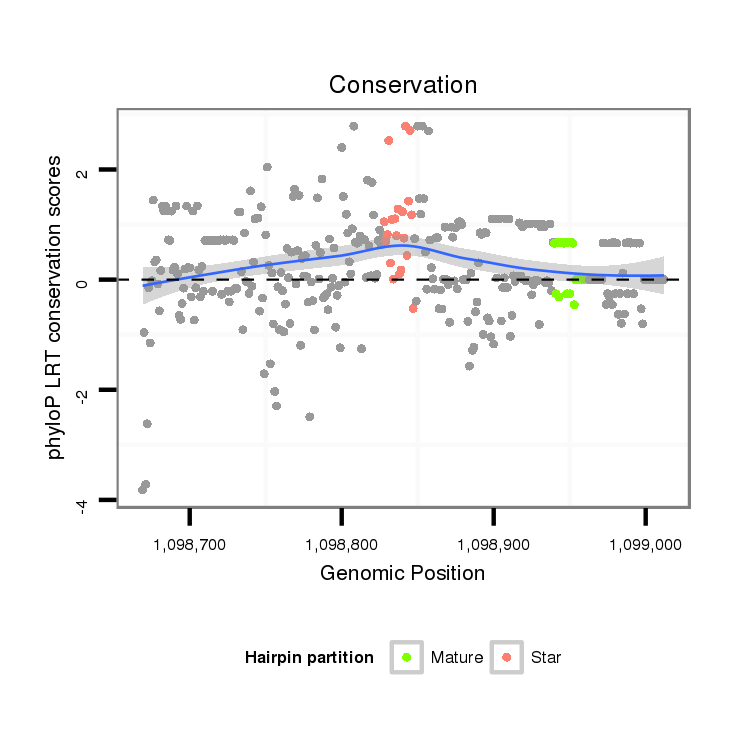
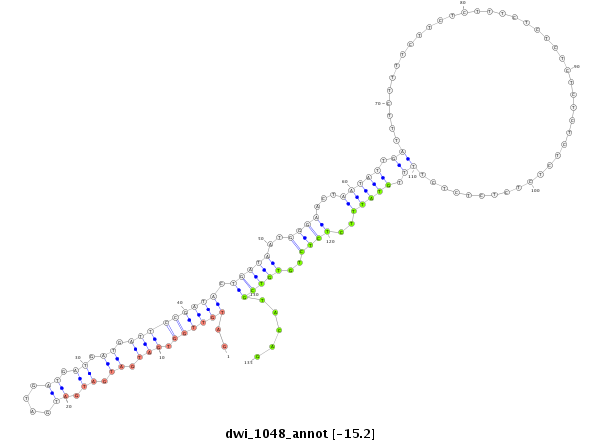
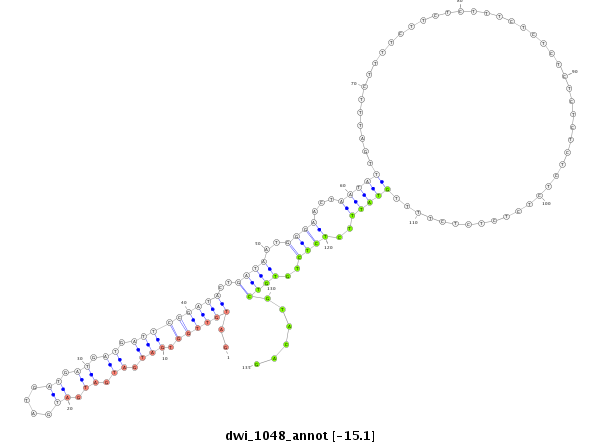
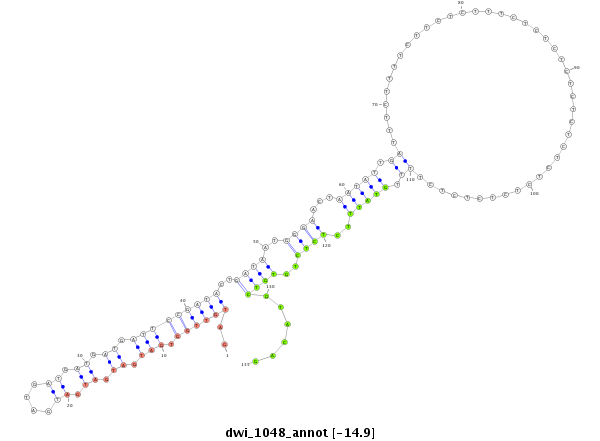
ID:dwi_1048 |
Coordinate:scf2_1100000004515:1098719-1098962 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -15.22 | -15.08 | -14.92 |
 |
 |
 |
CDS [Dwil\GK19992-cds]; exon [dwil_GLEANR_2592:1]; exon [dwil_GLEANR_2592:2]; CDS [Dwil\GK19992-cds]; intron [Dwil\GK19992-in]
| Name | Class | Family | Strand |
| (ATG)n | Simple_repeat | Simple_repeat | + |
| (TC)n | Simple_repeat | Simple_repeat | + |
| mature | star |
| --################################################----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ATATGCAATATAGTCTCTGGTTTGGGCTTTTCGCCGTAGTTGCCGCACCGGTAAGTGCCTCCCAATCCCCATCTCAATCTTAATCTCAATCCCAATCTCCTATCTCTATCTCTATGATTTTCCTAATGGATTGAAGATGATCATGATGATAATGATGTTGATGTTGGTGATGATGATGATGATGATGATGATGATTCCGATACTGATAATGGGAACTAATATTGATTTCTTTTCTTCTCTTTCTCTCTCTCTCTCTCTCTCTCTCTCTTTTGTATTTCTCTCTGTGTCGTACAGTTGGTGAGCGCTAACTATGGTCCAGGTGCTGGCGGACCAAGTGGTTCATA ***************************************************************************************************************************************************************..((((((.(((.((.((.((....)).)).)).))))))))).(((((..((((...(((((.................................................)))))..))))..))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
V118 embryo |
M020 head |
V119 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................TAGGGGATCGAAGATGATCA......................................................................................................................................................................................................... | 20 | 3 | 8 | 16.00 | 128 | 46 | 59 | 22 | 0 | 1 |
| ...........................................................................................................................TAGGGGATCGAAGATGATC.......................................................................................................................................................................................................... | 19 | 3 | 15 | 4.93 | 74 | 27 | 25 | 15 | 6 | 1 |
| ............................................................................................................................AGGGGATCGAAGATGATCA......................................................................................................................................................................................................... | 19 | 3 | 20 | 1.90 | 38 | 18 | 7 | 13 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................GCTGGCGGACCAAGTGGT..... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TAAGGGATCGAAGATGATC.......................................................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................GCACCGTTGGTGAGCGCTAACT.................................. | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................ATGGTCCAGGTGCTGGCGGACC............ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................GTATTTCTCTCTGTGTCGTACAG.................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......AATATAGTCTCTGGTTTGGGCT............................................................................................................................................................................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................ATGGTCCAGGTGCTGGCGGACCAAGTG....... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TACGGGATCGAAGATGATCA......................................................................................................................................................................................................... | 20 | 3 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................GGTTCAGGTCCTGGCGGA.............. | 18 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................AGGGTTGGGCTTTTCGCC..................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 4 | 0.50 | 2 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................TAGGGGATTGAAGATGATCA......................................................................................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................GATGTTGGTGATGATGATGA..................................................................................................................................................................... | 20 | 0 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 2 |
| ............................................................................................................................................................................ATGATGATGATGATGATGATGATTC................................................................................................................................................... | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................CTAGGGGATCGAAGATGATC.......................................................................................................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................TAGGGGATTGAAGATGATC.......................................................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................AGTGGATCGAAGATGATCA......................................................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................TGATGATGATGATGATGA............................................................................................................................................................... | 18 | 0 | 20 | 0.30 | 6 | 0 | 0 | 6 | 0 | 0 |
| ..........................................................................................................................TTAGTGGATCGAAGATGATC.......................................................................................................................................................................................................... | 20 | 3 | 7 | 0.29 | 2 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................TTAGGGGATTGAAGATGATCA......................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................TTAGGGATCGAAGATGATCA......................................................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TACGGGATCGAAGATGATC.......................................................................................................................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................AGGGATCGAAGATGATCA......................................................................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................GATGATGATGATGATGATGATT.................................................................................................................................................... | 22 | 0 | 20 | 0.10 | 2 | 0 | 0 | 1 | 0 | 1 |
| ................AAGGTTTGGGCTGTTCGCC..................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| .............................................................................................................................GGGGATTGAAGATGATCA......................................................................................................................................................................................................... | 18 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................TGCAGCACCGGTAATTGC.............................................................................................................................................................................................................................................................................................. | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................GATCTTGACGATAATGTTGTTG........................................................................................................................................................................................ | 22 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................GGGGATCGAAGATGATCAT........................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................ATGATGATGATGATGATGATT.................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................TATGGGATCGAAGATGATC.......................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................AGGGTTGGGCTATTCGCC..................................................................................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................ATGATGTTCATGTAGGTG............................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GATGATGATGATGATGATG.......................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
TATACGTTATATCAGAGACCAAACCCGAAAAGCGGCATCAACGGCGTGGCCATTCACGGAGGGTTAGGGGTAGAGTTAGAATTAGAGTTAGGGTTAGAGGATAGAGATAGAGATACTAAAAGGATTACCTAACTTCTACTAGTACTACTATTACTACAACTACAACCACTACTACTACTACTACTACTACTACTAAGGCTATGACTATTACCCTTGATTATAACTAAAGAAAAGAAGAGAAAGAGAGAGAGAGAGAGAGAGAGAGAGAAAACATAAAGAGAGACACAGCATGTCAACCACTCGCGATTGATACCAGGTCCACGACCGCCTGGTTCACCAAGTAT
**************************************************..((((((.(((.((.((.((....)).)).)).))))))))).(((((..((((...(((((.................................................)))))..))))..))))).....*************************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M045 female body |
V117 male body |
V119 head |
V118 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ................GACCAAACCCGACAAGGGGCAA.................................................................................................................................................................................................................................................................................................................. | 22 | 3 | 1 | 81.00 | 81 | 72 | 7 | 2 | 0 |
| ................GACCAAACCCGACAAGGGG..................................................................................................................................................................................................................................................................................................................... | 19 | 2 | 1 | 49.00 | 49 | 49 | 0 | 0 | 0 |
| ................GACCAAACCCGACAAGGGGCA................................................................................................................................................................................................................................................................................................................... | 21 | 2 | 1 | 47.00 | 47 | 42 | 4 | 1 | 0 |
| ................GACCAAACCCGACAAGGGGC.................................................................................................................................................................................................................................................................................................................... | 20 | 2 | 1 | 28.00 | 28 | 26 | 1 | 1 | 0 |
| .................ACCAAACCCGACAAGGGGCAA.................................................................................................................................................................................................................................................................................................................. | 21 | 3 | 3 | 16.33 | 49 | 43 | 4 | 2 | 0 |
| .................ACCAAACCCGACAAGGGGCA................................................................................................................................................................................................................................................................................................................... | 20 | 2 | 1 | 16.00 | 16 | 16 | 0 | 0 | 0 |
| .................ACCAAACCCGACAAGGGGC.................................................................................................................................................................................................................................................................................................................... | 19 | 2 | 2 | 9.50 | 19 | 19 | 0 | 0 | 0 |
| ..............GAAACCAAACCCGACAAGGGGC.................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 1 | 9.00 | 9 | 9 | 0 | 0 | 0 |
| ..............GAAACCAAACCCGACAAGGGGCA................................................................................................................................................................................................................................................................................................................... | 23 | 3 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 |
| ..................CCAAACCCGACAAGGGGCA................................................................................................................................................................................................................................................................................................................... | 19 | 2 | 3 | 7.00 | 21 | 21 | 0 | 0 | 0 |
| ...............AAACCAAACCCGACAAGGGGCA................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 4 | 6.50 | 26 | 22 | 3 | 1 | 0 |
| ...............GGACCAAACCCGACAAGGGGCA................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 |
| ................AACCAAACCCGACAAGGGGCA................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 7 | 3.43 | 24 | 19 | 5 | 0 | 0 |
| ...............AAACCAAACCCGACAAGGGGC.................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 6 | 2.50 | 15 | 13 | 0 | 2 | 0 |
| ...............GGACCAAACCCGACAAGGGG..................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 5 | 2.20 | 11 | 11 | 0 | 0 | 0 |
| ..............GAAACCAAACCCGACAAG........................................................................................................................................................................................................................................................................................................................ | 18 | 2 | 20 | 1.65 | 33 | 33 | 0 | 0 | 0 |
| ..............GAAACCAAACCCGACAAGGGG..................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 5 | 1.60 | 8 | 8 | 0 | 0 | 0 |
| ................................TGGCATCAACGGCGTGGCCTAT.................................................................................................................................................................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...............AGACCAAACCCGACAAGGGGCA................................................................................................................................................................................................................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................GACCGCCTGGTTCACCAAGTA. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................TGGTGGCATCAACGGCGTGGCC..................................................................................................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................GACCAAACCCGACAAGGG...................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 6 | 1.00 | 6 | 6 | 0 | 0 | 0 |
| ................GACCAAACCCGACAAGTGGC.................................................................................................................................................................................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................AACCAAACCCGACAAGGGGC.................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 14 | 0.86 | 12 | 12 | 0 | 0 | 0 |
| ................GACCAAACCCCACAAGGGGCA................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................ACCAAACCCGACAAGGGG..................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 7 | 0.43 | 3 | 3 | 0 | 0 | 0 |
| ................AACCAAACCCGACAAGCGGCAA.................................................................................................................................................................................................................................................................................................................. | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ................GACCAAATCCGACAAGGGGC.................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ..............GAAACCAAACCCGACAAGGG...................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................GACCGCCGGGTTGACCATGT.. | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ..............AAGACCAAACCCGACAAGGGG..................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ..............GGGACCAAACCCGACAAG........................................................................................................................................................................................................................................................................................................................ | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ..................CCAAACCCGACAAGGGGC.................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| .......................................................TCTGAGGGTTAGGGGTAG............................................................................................................................................................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................................AGAAAGAGAGAGAGAGAGA........................................................................................ | 19 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 |
| ..............AGGACCAAACCCGACAAGC....................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................CGTGATGATAACTCAAGAAA................................................................................................................ | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 |
| .........................................................GCAGGGTTACGGGTAGGGTTA.......................................................................................................................................................................................................................................................................... | 21 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| .................ACCAAACCCGAAAAGGGCAA................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................ACCACTACTACTACTACTACTACTA........................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ........................................................................................GAGGGTTAGAGGATAGGGTT............................................................................................................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droWil2 | scf2_1100000004515:1098669-1099012 + | dwi_1048 | ATATGCAATATAGTCTCTGGTTTGGGCTTTTCGCCGTAGTTGCCGCACCGGTAAGTGCCTCCCAATCCCCATCTC----------------------------AATCTTAATCTC-------------------------------------------------------------------------------AATCCCAATCTC-CT--------------A----TCTCTAT-CTCTATGATT--TTCCTA----------------ATGGATT-----------------------------------GAAG----------ATGATCATGATGAT--AATGATG----TTGA-----------------------------TGTTGGTG---ATGATGA------------TGATGATGATGATGATGATTCC---------------------------GATACTGATAATGGGAAC----------------------------------TA--------------------------------ATATTGATTTCTTTTCTTCTCTTTCT------------C-TCTCTCTCTCTCTCTCTCTCTCTTTTG----TATTTCTCTCTGTGTCGTACAGTTGGTGAGC--------------------------------------------------------------------------------------------------------------------------------------GC----TAACTATGGTCCAGGTGCTGGCGGACCAAGTGGTTCATA |
| droVir3 | scaffold_13042:2972544-2972707 + | CTAATG-------------------------------------------------------CTAATGCTAATGC----------------------------------TAATGCT----------------------------------------------------------------------AATGCTAA----------------TGCTAATGCT----------------------------------AATGCTAATGCTAATGCT---AATGCTAATGA-TAATGATAATGA----------------TAATGATAA----TGA------TAAT--GATAAT-G---ATAATG---------ATAATGATAA--TGATAATGATAATG------ATAA------------TGATAATGATAAT------------------------------------------GATA------ATG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT-------------- | |
| droMoj3 | scaffold_6308:564219-564388 - | CTAATG-------------------------------------------------------CTAATACTATTAC----------------------------------TAATATT----------------------------------------------------------------------AATGCTAAAGTTAT----------TACTAATTCT----------------------------------AATGGTAATGATAATGAT---AATGATAATAA-TAATGATAATGA----------------TAATGATAA----TGA------TAAT--GATAAT-G---ATAATG---------ATAATGATAA--TGATAATGATAATG------ATAA------------TGATAATGATAAT------------------------------------------GATA------ATG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT-------------- | |
| droGri2 | scaffold_15245:14166035-14166220 - | ATGA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG----------GCGA---TCA-----------TG----ATGA--------------------------TGATGA---TGATGAAGATGATGATGATGATGATGATGATGATGATGAT------------------------------------------G------ACA-------------------------------ATTG--------------------------------TACG---------------------------------------------------------------------------------------------------------------------------------------------GAACCGTCACTCGCTTCCACTTCCGTTCGGCTTTTATTG-------------TTTTCGCACCTCGCCTTCCGTTCGTTTGGCGCTCGGGTCGCGATCGGTCGACGCCATCGATGCCGGCGGA-------------- | |
| dp5 | XL_group1a:4033727-4033860 - | ATACTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCTCTATCT--------------A----TCGCACT-CACTGTGTTT--TGC--C----------------TTTTGTT----------GATGATAATGA----------------TGATGGTTA----TGA------TGAT--GATGTT-A---TTGATG---------ATAACGATGA--TGATGATGATGATG------ATGA------------TGATGATGATGGT------------------------------------------GATGATG---G-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_1:7860187-7860387 + | GA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGATCATAA-----------TG----ATGA-----------------------TGATGATGATGATG------ATGA------------TGATGATGATGATGAT------------------------------------------G------ATGTGTTTATTCCTATATAGTGAGGGGTTGGGGGCTCAGTACACACTCTCATATATGGCCTTCTCATGGTTCAT------CATGTCTTCGCTGTCTGTGAGTGTGTTTT-ACCCCCCCTCTGGCTCCCTCTCTTCTGGTCTCAATTCTTTTTGG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GT-------------- | |
| droAna3 | scaffold_12943:3579822-3579995 + | GGG----ATTCAGTCTC--GGATCCAGCTTCTTCG--------------------------------------------------CCTTGCAGGAT----------CCGGAACCGGATCCACCAGATCCTA---------------------------------------------------------ATC-CCAGGCCACCTCCA------------------------CCACC-CCCCG----T--CGC--C----------------GTCAGAT-----------------------------------GGTG-------------A---TGG-----------TG----ATGG--------------------------TGGTGG---TGGTGCTGCTGC------------TGCTGATGATGATGGT---------GATGCCGGT----GCGGAT-----------GCTG------CGA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droBip1 | scf7180000396696:571228-571370 - | TCTC------------------------------TATCTC-----------------------------TCTCTCTCTCTCTCTC----TCAACAC--ACATA---------------------------------CTACT-------------------------------------------------T-ACTC----TCTCTATCT--------------A----TCGCACT-CACTGTGTTTTTTGT--C----------------TTTTTGT----------GAT----------------------GA-------------TGA---TGA-----------TG----ATGA-----------------------TAATGATGATGATG------ATGA------------TTGCGATGATGATGAT------------------------------------------G------GCG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droKik1 | scf7180000302291:20901-21136 + | ACATGAAACTCGGTCTCTGGTTTGGCATCTTCGCCATCGCCGCCGCGCCGGTTAGTAATTCCCAAC------------------------------------------------------------TCCTAACACGCTGCCTCCTTCCTCTGTAATTTCCGACTAAATATTTCGCGGCGGAGGGATACGTG-TAACTCGCAACTAATCT--------------CTTTGCC--ACCTTCTTG----T--CTC--C----------------CTCAGCT-----------------------------------GGTG-------------A------G-----------------CGCTA---------ACTATGG--------CCATGGGCACG------GTCA------------TGGCCACCACAGTGGT------------------------------------------G------GTG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------GACAGTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453826:683503-683625 - | CTCAAC----------------------------------------------------------------------------------------------------CTCAATTTG-----------------------------------------------------------------------------GACCACC----------ATCTCT--C-----C----TCCTCCT-TCCCATGCTT--GTTGTT-----GTTGGTATTGCTCCTGTT-----------------------------------GA-------------TGA---TCA-----------TG----ACGA-----------------------CGATGGTGATGATG------ATGA------------TGATGATGATGTT------------------------------------------GCTC------CTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEle1 | scf7180000491044:892869-893013 - | TTAGGT-------------------------------------------------------TG----------------------------------------------AATTTC----------------------------------------------------------------------AATTT------CGC----------AGCTAT--CT-----------------CTTTT-----------------------------------TGCTACTTAC-TT-------------------------TG-------------A------TTACACAA---TG----ATGA-----------------------TGATGTTGATGATG------ATGA------------TGATGATGATGATGAA------------------------------------------A------GCT----------------------------------C-----------------------C--------ACTTTGCTTCATTATTTTTTTTTTGT------------T-TCCCC--------------------------------------------------------------------------------------------------------------------------------------------------------ATCTCC-------------------------------------------------ATC-------------------------- | |
| droRho1 | scf7180000769923:3999-4163 + | ATCA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAT-----------------------------------GATG-------------A------TGATAAGA---GG----ATGA-----------------------------CGATGACG------ATGA------------TGATGATGATGATGAT------------GACGTTCCTTGCAGAGGATACAACCCTCAT-----------------------------------------------------------GCAACGTTGT-----TGTTCATTGATTTTCTTTTTTATTTTTTG------------TGTTCCCCCTTTTGGTTTGCTCCTCTT----------------------------------------------------------------------------------------------------------------------------------------------------TCTTTTCCACCC----------------------------------------AG-------------- | |
| droBia1 | scf7180000301709:259300-259458 - | ATGA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG----------ATGA---TGA-----------TG----ATGA--------------------------TGATGA---TGATGATGATGA---------------TGATGATGA---G---ATGGACATTGAGTTT---------------------GAGG------ACG-------------------------------TTTG--------------------------------ATAT---------------------------------------------------------------------------------------------------------------------------------------------GGTCCGTTTTTTTTTAATCTTACCAACTAGCTCTTACAATGTAGAACCAACTTTTCCTTTCTTCATTTTCTTCTCTATTG----------------------------------------TA-------------- | |
| droTak1 | scf7180000415399:908217-908391 - | GCGGGTGATTT--------------------------------------------------C----------------------------------------------CAATATT----------------------------------------------------------------------ATTCCC-AAGA----TATTGATCTTTCGAATTCC------------------AATAGGTCTTCTGA--T--------TTTTACATT---GATGATGATGA-TGATGATGATGATGATGATGATGATG-----------ATGATGA-----------------T-GATGATGATG---------ATGATGAT--GATGATGATGATGATG------ATGA------------TGATGATGATGAT------------------------------------------GATG------ATG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEug1 | scf7180000409040:73998-74208 + | CATC---------------------------------------------------------TG----------------------------------------AATCTGAATCTG----------------------------------------------------------------------AATGTG-AAGTTGC----------AACTGT--CGAATG-------------TTCGAT-----------------------------------GATGAT----------------------CG--C----TG-------ATGACTA---AAA-----------TG----ATGATCATAATAACCATAATGA-----------TGA---TG------ATGA------------TGATGATGATGGT------------------------------------------------------G----------------------------------C-----------------------T--------G-----------------------------------------------------------------------------------------------------GATGATGGCGACGTTATTGTGCCAAGGTAAGAGATCGTTGCCAAATTTGTTTTTCTTTTTCGTTTTTTTTTGTTTTTTTTTG-------------GTTTTC-------------------------------------------------TTT-------------------------- | |
| dm3 | chrX:5123935-5124130 + | ATATGCGATTTCATCTC--GTTTGTTGCTTTCTCTGTCTC-----------------------------TATCTCTCTCTCTCTCTCTTTCAACACTCACATAAA----------TATA------------------------------------------------------------TATATATATATA-CAAATCCAACTCTATCT--------------A----TCGCACT-CACTGTGTTT--TGC--C----------------TTTTGTT-----------------------------------GATG-------------A------TGATTTGA---TG----ATGA------------------T--AATAATGATGATGATG------ATGA------------TGGTTATGATGATAAT------------------------------------------G------GCG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droSim2 | x:4772266-4772443 + | ATATGCGATTTCATCTC--GTTTGTTGCTTTCTCTGTCTC-----------------------------TATCTCTCTCTCTCTCTC--TCCACACTCACATAAA----------TAT----------------------------------------------------------------------ATA-TAAATCCAACTCTATCT--------------A----TCGCACT-CACTGTGTTT--TGC--C----------------TTTTGTT-----------------------------------GATG-------------A------TGATTTGA---TG----ATGA--------------------------TAATGATGATG------ATGA------------TGGTTATGATGATGAT------------------------------------------G------GCG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droSec2 | scaffold_21:464118-464244 + | ATTT---------------------------------------------------------CG----------------------------------------AAACCAAAC----------------------------------------------------------------------------------------------------TCT--C-----------------------------------------------CACTTT---GTTGATAATTAC-CATGACAATGATGATGATGA--ATG-----------ATGACGA---TAA-----------CG----ATGA-----------------------CGATGTCGATGATA------ATGA------------TGATGATGATGA---CAATT---------------------------ACAACCGTGATGATG---G-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | X:14297714-14297855 + | CATC---------------------------------------------------------TG----------------------------------------AATCCGAATCTG-----------------------------------------------------------------------------AAGTTGC----------AACTGT--CGAATG-----------T-TTCGAT-----------------------------------GATGATCGCCGATGACGCTGA---TGACTATAATG-----------ATGATGA------TGA--------TA----ATGATG---------ATGATGAT--GACAATGGTGATAATG------ATGA------------TGATGATGATGGT------------------------------------------GGTG------GTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEre2 | scaffold_4690:2478928-2479102 + | ATATGCGATTTCGTCTC--GTTTGTTGCTTTCTCTGTCTC-----------------------------TATCTCTCTCTTTCTC----TCCACACTTACATTAA----------TAT--------------------------------------------------------------------ATATA-TAAATCCAACTCTATCT--------------A----TCGCACT-CACTGTGTTT--TGC--C----------------TTTTGTT-----------------------------------GATG-------------A------TGATTTGA---TG----A-----------------------------TAATGATGATG------ATGA------------TGGTTATGATGATGAT------------------------------------------G------GCG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 12:33 AM