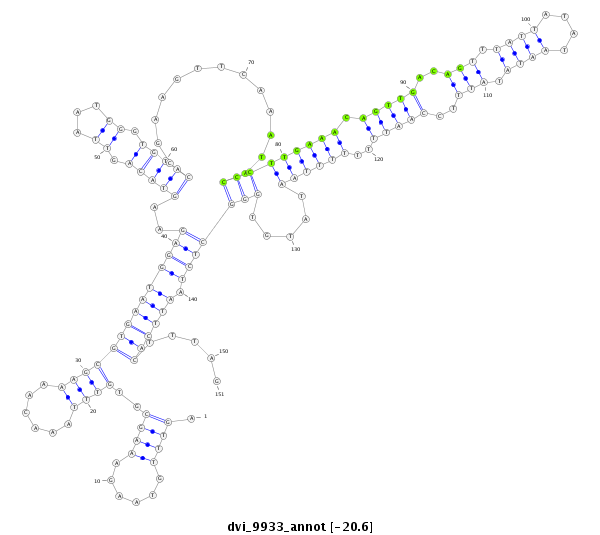
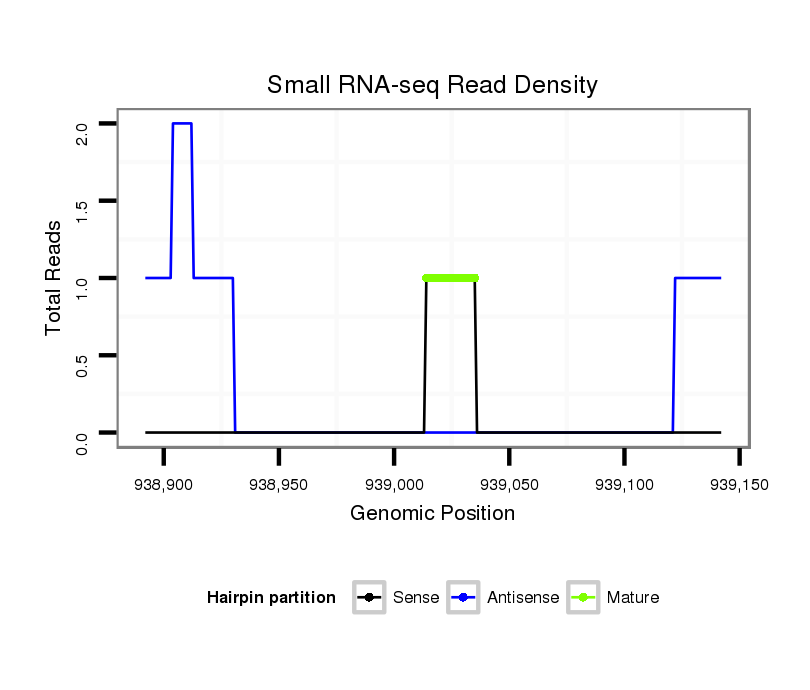
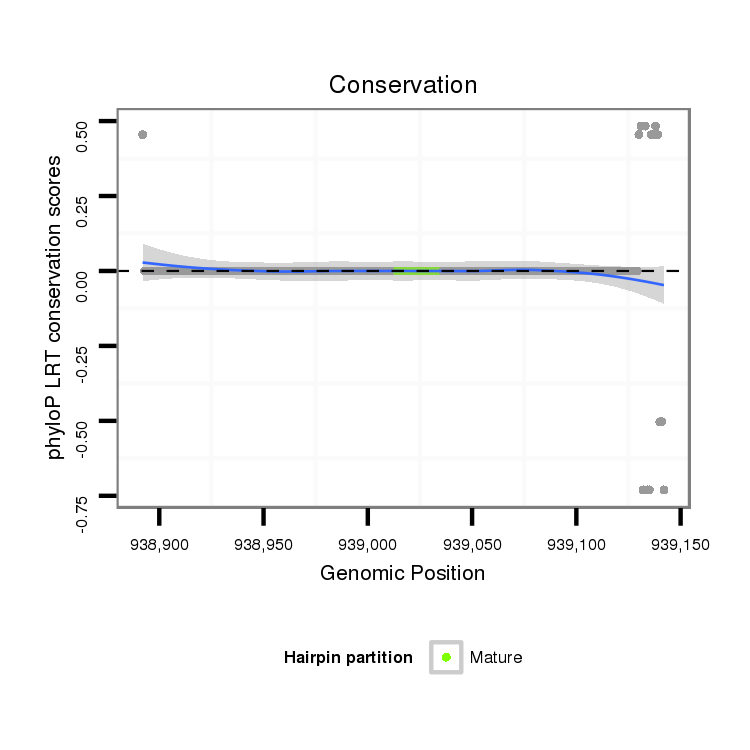
ID:dvi_9933 |
Coordinate:scaffold_12963:938942-939092 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.6 | -20.4 | -20.4 |
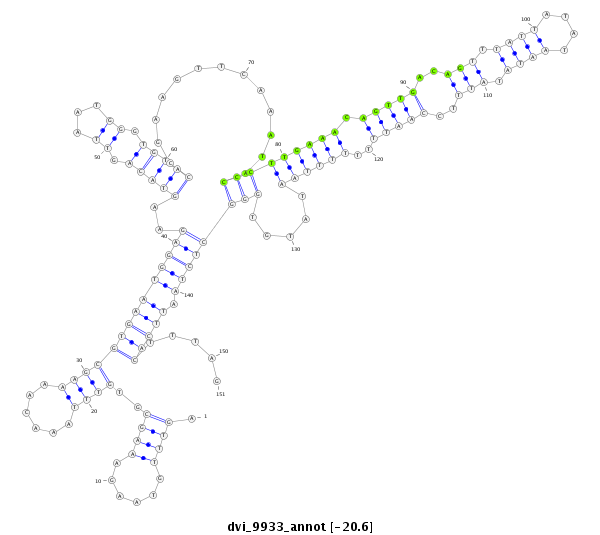 |
 |
 |
CDS [Dvir\GJ14585-cds]; exon [dvir_GLEANR_1454:2]; intron [Dvir\GJ14585-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CACGCTTTAATTGCACATGCGTGCGCTGCCCACAATGTAAACGATTACGCAGTTTGTAAGAAAGCGTGTTTAAACAAAAGCGTGAATGGAGAAGTACAGTTAATGGGTGTACCGAAGTTCAAATACCCTTGAAACAGTTGACAGTTTATTATATAATATATTTCCAATTTTTTTTAATATGTGGGCTCTAATTCACTTTAGGGTCAACCTGCGCAAACGTCTGCCCCGCGGCAGCCGCCAGTTTGCACTGA **************************************************.((((......))))..((((......))))((((((((((..(((((.((...)).))))).............(((((((((.(((((..(((.((((....)))).)))..)))))..)))))).....))))))).)))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060658 140_ovaries_total |
SRR060678 9x140_testes_total |
SRR060683 160_testes_total |
M027 male body |
SRR060666 160_males_carcasses_total |
M028 head |
M061 embryo |
SRR060684 140x9_0-2h_embryos_total |
V047 embryo |
SRR060667 160_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060679 140x9_testes_total |
SRR060687 9_0-2h_embryos_total |
GSM1528803 follicle cells |
V053 head |
SRR060659 Argentina_testes_total |
SRR060672 9x160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................TGTAAACGATAACGCAGAT..................................................................................................................................................................................................... | 19 | 2 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................GTAAACGATAACGCAGAT..................................................................................................................................................................................................... | 18 | 2 | 4 | 4.50 | 18 | 3 | 0 | 0 | 0 | 3 | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................ATACCCTTGAAACAGTTGACAGTC......................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................AAGACCGTGAATGGAGGAGT............................................................................................................................................................ | 20 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................ATACCCTTGAAACAGTTGACAG........................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GGTGAACCTGCGTAAACCTC.............................. | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TGTAAACGATAACGCAGA...................................................................................................................................................................................................... | 18 | 2 | 20 | 0.15 | 3 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................GCCCATGATGTAAACGAT.............................................................................................................................................................................................................. | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................TAAGGAATCGTGTTTAAAAAAA............................................................................................................................................................................. | 22 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CAATCTAAACGGTTACGAAG....................................................................................................................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................TGTAAACGATAACGCAGATG.................................................................................................................................................................................................... | 20 | 3 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................CACTTTAAGGACAACCTGT....................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TTTAAACGATAACGCAGAT..................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................GTTAAACGATAACGCAGTT..................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................ATTACGAAGTTTGGAGGAA............................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTGCGAAATTAACGTGTACGCACGCGACGGGTGTTACATTTGCTAATGCGTCAAACATTCTTTCGCACAAATTTGTTTTCGCACTTACCTCTTCATGTCAATTACCCACATGGCTTCAAGTTTATGGGAACTTTGTCAACTGTCAAATAATATATTATATAAAGGTTAAAAAAAATTATACACCCGAGATTAAGTGAAATCCCAGTTGGACGCGTTTGCAGACGGGGCGCCGTCGGCGGTCAAACGTGACT
**************************************************.((((......))))..((((......))))((((((((((..(((((.((...)).))))).............(((((((((.(((((..(((.((((....)))).)))..)))))..)))))).....))))))).)))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060678 9x140_testes_total |
M061 embryo |
SRR060681 Argx9_testes_total |
SRR060675 140x9_ovaries_total |
V053 head |
SRR060685 9xArg_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................TAATTGAAGTCCCAGTTG........................................... | 18 | 2 | 5 | 1.40 | 7 | 0 | 1 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TAATTGAAGTCCCAGTTGAAC........................................ | 21 | 3 | 3 | 1.33 | 4 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................CGTCGGCGGTCAAACGTGACT | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GTGCGAAATTAACGTGTACGC...................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............CGTGTACGCACGCGACGGGTGTTACAT.................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................GCGTTTGCAGGCGGGGCGCCG................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................GTTACATTTGCTACTGCGC........................................................................................................................................................................................................ | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TAATTGAATTCCCAGTTGAAC........................................ | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................ACCCGAGATTAAGTTCAAT................................................... | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................AAAAATATATTCTACAAAGGTT.................................................................................... | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TTGCATAAGGGGCGCCGTA................. | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TAGGACGTTTGTCAACTGTC........................................................................................................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................GTTGCAATTGCTAATGGGT........................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TTGCACAAGGGGCGCCGTA................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:938892-939142 + | dvi_9933 | CACGCTTTAATTGCACATGCGTGCGCTGCCCACAATGTAAACGATTACGCAGTTTGTAAGAAAGCGTGTTTAAACAAAAGCGTGAATGGAGAAGTACAGTTAATGGGTGTACCGAAGTTCAAATACCCTTGAAACAGTTGACAGTTTATTATATAATATATTTCCAATTTTTTTTAATATGTGGGCTCTAATTCACTTTAGGGTCAACCTGCGCAAACGTCTGCCCCGCGGCAGCCGCCAGTTTGCACTGA |
| droMoj3 | scaffold_6500:27655760-27655773 + | C---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATTGGGCACCAC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/17/2015 at 05:01 AM