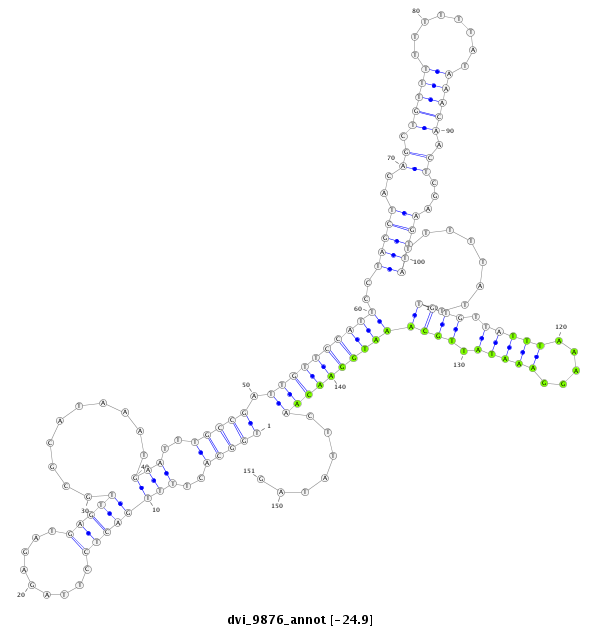
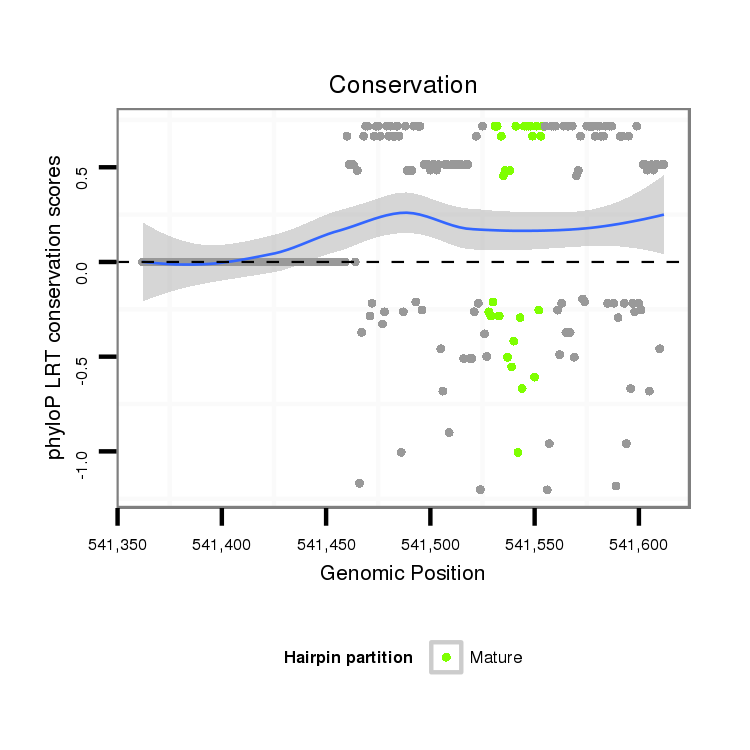
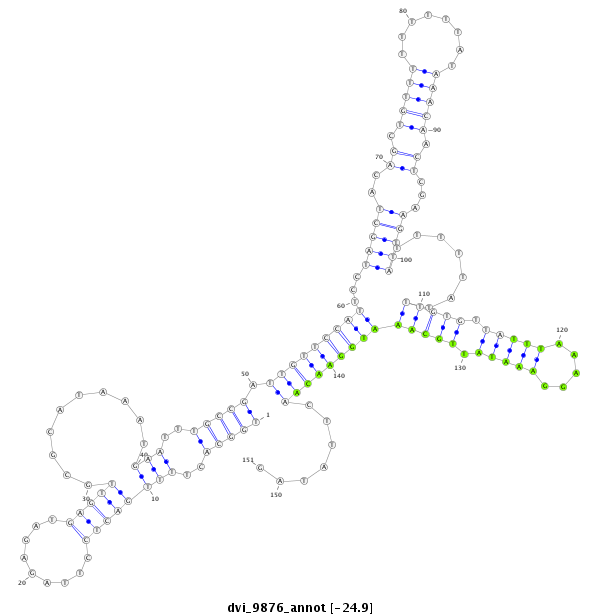
ID:dvi_9876 |
Coordinate:scaffold_12963:541412-541562 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -24.9 | -24.7 | -24.7 |
 |
 |
 |
exon [dvir_GLEANR_1439:5]; CDS [Dvir\GJ14439-cds]; intron [Dvir\GJ14439-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CAATAAAGAAAACATGTCATTCAAATAGTTCTTATTGCGCTCCTTAGAGATGGCACTTTTGACTCCTTAGAGATGAGTTGCGCATAAATGAATTTGCCGATTGTTCCATTCCTAGCTACAGCTGTTTTTTTTTATAAACAACTCGAAGTTATTTTTATTTGTGTTATTTAAAGGAAATATTGCAAATGGAACAACTTATAGATGTGAGCTGCATTAAGAAGGAGTACACGGAAGCTAACAATGCAGATAAA **************************************************(((((..((((((((.........)))))..........)))..))))).((((((((((..(((((..((.(((((........))))).))...)))))........((((.(((((.....))))).)))))))))))))).......************************************************** |
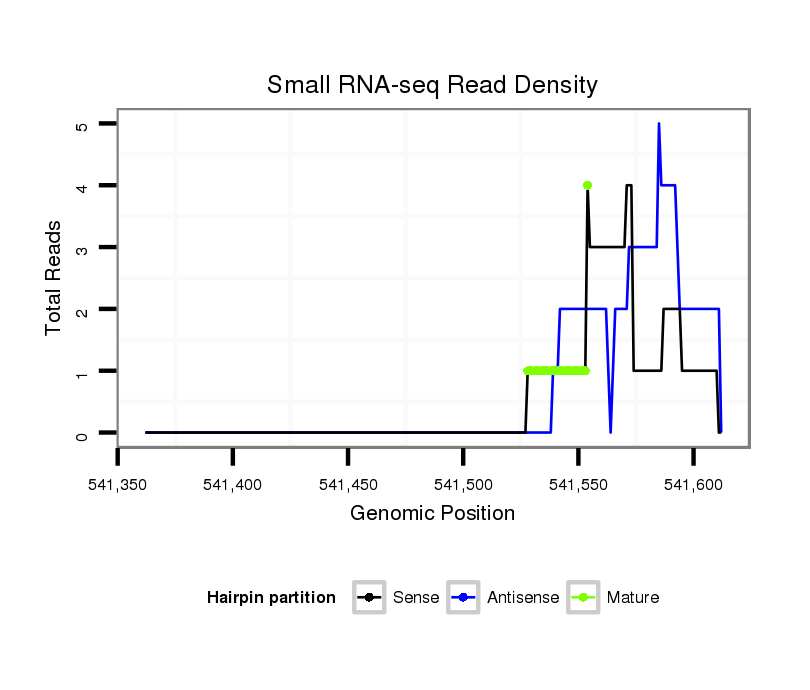
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060678 9x140_testes_total |
SRR060681 Argx9_testes_total |
M047 female body |
SRR060665 9_females_carcasses_total |
SRR060689 160x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................AACTTATAGATGTGAGCTGC....................................... | 20 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................GTGCTGCATTAAGAAGGAGTA......................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CTTAGAGATGAGTAGCGCATA..................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TGCATTAAGAAGGAGTACACGGAA.................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................TTTAAAGGAAATATTGCAAATGGAACA.......................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TATAGATGTGTGCTGCATTAAGAAGG............................. | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................ACACGGAAGCTAACAATGCAGATA.. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TGAATTGGCCAATTGTTCC................................................................................................................................................ | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................TATTTAGAGGAAGTATTG..................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................CGAAGCTAAGAATGCAGA.... | 18 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................GGCGATCCTAAGAGATGGC..................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................AAATGGTCCTTATTACGCT.................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
GTTATTTCTTTTGTACAGTAAGTTTATCAAGAATAACGCGAGGAATCTCTACCGTGAAAACTGAGGAATCTCTACTCAACGCGTATTTACTTAAACGGCTAACAAGGTAAGGATCGATGTCGACAAAAAAAAATATTTGTTGAGCTTCAATAAAAATAAACACAATAAATTTCCTTTATAACGTTTACCTTGTTGAATATCTACACTCGACGTAATTCTTCCTCATGTGCCTTCGATTGTTACGTCTATTT
**************************************************(((((..((((((((.........)))))..........)))..))))).((((((((((..(((((..((.(((((........))))).))...)))))........((((.(((((.....))))).)))))))))))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060681 Argx9_testes_total |
SRR060670 9_testes_total |
SRR060679 140x9_testes_total |
SRR060680 9xArg_testes_total |
V047 embryo |
V116 male body |
SRR060684 140x9_0-2h_embryos_total |
SRR1106723 embryo_12-14h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................CATGTGCCTTCGATTGTTACGTCTATT. | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................ACGTTTACCTTGTTGAATATC.................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CACTCGACGTAATTCTTCCTC........................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................CGTAATTCTTCCTCATGTGCC.................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................AGGATCGATGTCGACAAAAAAAATATT................................................................................................................... | 27 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................ACTCGACGTAATTCTTCCTCATGTGCCT................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................ATAACGTTTACCTTGTTGAATATCT................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TCTCTGCCGTGAAAACTGAGGAATCT.................................................................................................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........TGTACTGTAGGTTTATCAAG............................................................................................................................................................................................................................ | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................AGGTATGCATCGATGTCG................................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................CTAATGCGAGAAATCTCTA........................................................................................................................................................................................................ | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................TCGGTGTCAACAAAAAAAACTA.................................................................................................................... | 22 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:541362-541612 + | dvi_9876 | CAATAAAGAAAACATGTCATTCAAATAGTTCTTATTGCGCTCCTTAGAGATGGCACTTTTGACTCCTTAGAGATGAGTTGCGCATAAATGAATTTGCCGATTGTTCCATTCCTAGCTACAGCTGTT------------------TTTTTTTATAAACAACTCGAAG------------------------TTATTTTTATTTGTGTTATTTA--------AAGGAAAT-------ATTGCAAATGGAACAACTTATAGATGTGAGCTGCATTAAGAAGGAGTACACGGAAGCTAACAATGCAGATAAA |
| droMoj3 | scaffold_6500:12044332-12044470 + | --------------------------------------------------------------------------------------------------G----TCTCATCTCTAGAAACAGCTGGAATAAACAAGTTTGTTTTTTTTTTTTAC------------------------------------------------AGTATTCACTA--------AGGGAGAAACAATTTTTGGAAAATGCAGCAATATATGGATATAAACTGCATTAAGAAGGAATATCCGGATGATTATG----------- | |
| droGri2 | scaffold_15126:3080141-3080312 - | --------------------------------------------------------------------------------------------------GATT--GTCATCCCTAGCTACAGCTGAT------------------T---TCTATAAACAACTTTAACCACATTCATAAGATACTCTTTGGTTTATTTATAAATGACTGATCCAAATGACTGAGG------------TTATAAAATGGAACAAGCTATACTTATAA---GTGTTAAGAAGGAGTACGAGGTGGAAAACAATGAAGATGAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/17/2015 at 04:37 AM