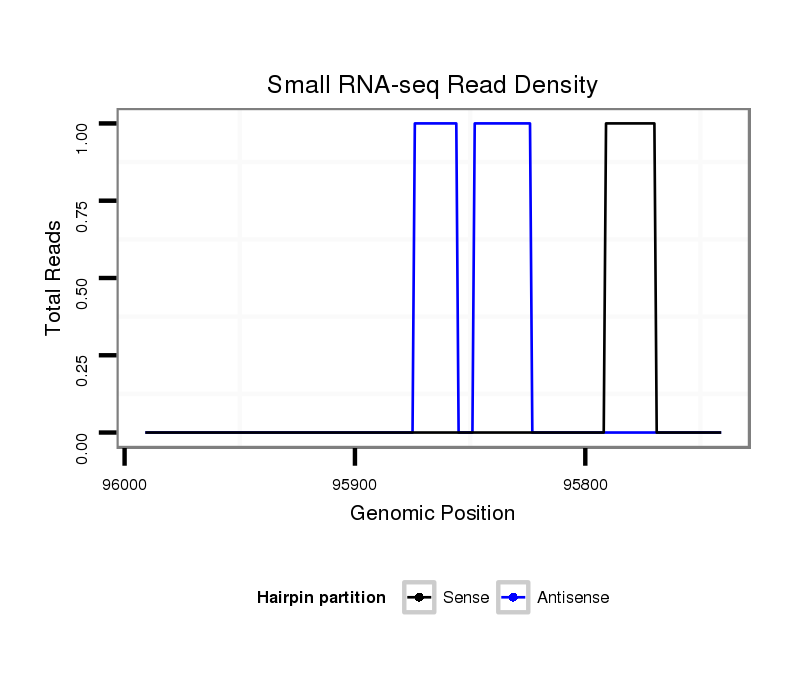
ID:dvi_9823 |
Coordinate:scaffold_12963:95791-95941 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
No conservation details. |
CDS [Dvir\GJ14156-cds]; exon [dvir_GLEANR_1411:1]; intron [Dvir\GJ14156-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTATCTTGGCACCGCAGTCCTTTTGTGAGACTAAACCAATTATAAGTCTGGTGAGAAAATTAGAATATTTTGATGCACTACAGTACAGTTTAGAATATATAGAGTATTATCTGATTTAGCTTCCAGGGCAAAACATGTTTTAATCAGGTCTGCCAACCCAACAATATCTGTCGCTCATGAGCCCCATTAATTAATTAAGCACTTTAATTTCTAATTGACCGCTTTACGGTTTATAACATATATACGTCGTT **************************************************..(((.(((((((((((((((((((..(((..........)))..))).))))))))..))))))))..)))...((((....((((.......(((((.((.........))..))))).....)))).)))).................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060675 140x9_ovaries_total |
SRR060689 160x9_testes_total |
V053 head |
SRR060679 140x9_testes_total |
M047 female body |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................ACTTTAATTTCTAATTGACCGC............................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................CAAATTTTAAGTCTGTTGAGAA.................................................................................................................................................................................................. | 22 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................TAAGACTGGTGAGAAAAA............................................................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......TGGCACCGCGTTCCTCTTGT................................................................................................................................................................................................................................. | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................TGAGAAAATTAGAGTGTT...................................................................................................................................................................................... | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................TTATAAGTCTGGTGTAAAA................................................................................................................................................................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................GAAGTCTGGTGAGAAAGTA.............................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
|
CATAGAACCGTGGCGTCAGGAAAACACTCTGATTTGGTTAATATTCAGACCACTCTTTTAATCTTATAAAACTACGTGATGTCATGTCAAATCTTATATATCTCATAATAGACTAAATCGAAGGTCCCGTTTTGTACAAAATTAGTCCAGACGGTTGGGTTGTTATAGACAGCGAGTACTCGGGGTAATTAATTAATTCGTGAAATTAAAGATTAACTGGCGAAATGCCAAATATTGTATATATGCAGCAA
**************************************************..(((.(((((((((((((((((((..(((..........)))..))).))))))))..))))))))..)))...((((....((((.......(((((.((.........))..))))).....)))).)))).................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
SRR060678 9x140_testes_total |
SRR060655 9x160_testes_total |
SRR060658 140_ovaries_total |
SRR060673 9_ovaries_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060657 140_testes_total |
SRR060659 Argentina_testes_total |
SRR060676 9xArg_ovaries_total |
SRR060679 140x9_testes_total |
M061 embryo |
SRR060683 160_testes_total |
SRR060666 160_males_carcasses_total |
M047 female body |
SRR060674 9x140_ovaries_total |
SRR060689 160x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................TTCATTAATTCGTGAGATTAA.......................................... | 21 | 2 | 2 | 2.00 | 4 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TTCATTAATTCGTGAGATTAAACA....................................... | 24 | 3 | 2 | 2.00 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CATTAATTCGTGAGATTAAACA....................................... | 22 | 3 | 4 | 1.50 | 6 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CATTAATTCGTGAGATTAA.......................................... | 19 | 2 | 5 | 1.40 | 7 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................AGTCCAGACGGTTGGGTTGTTATAG................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TCATTAATTCGTGAGATTAAA......................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TCGAAGGTCCCGTTTTGTA................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TTCATTAATTCGTGAGATTA........................................... | 20 | 2 | 3 | 0.67 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................ATTAATTCGTGAGATTAA.......................................... | 18 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................CAAATAGTGTAGGTATGCAGC.. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CATTAATTCGTGAGATTAAA......................................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AGAACTGTGGTTTCAGGAAAA................................................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CATTAATTCGTGAGATTAAAC........................................ | 21 | 3 | 13 | 0.15 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................CAAATAGTGTATGTATGCA.... | 19 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................TGTACCAAAGTAGTCCAGG.................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AATTCGTGAGATTAAAGA....................................... | 18 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................AAATAGTGTAGATATGCATC.. | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................AATTCGTGAAATTAAACA....................................... | 18 | 1 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TCGAAGGTCCCCCTTTGC.................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12963:95741-95991 - | dvi_9823 | GTATCTTGGCACCGCAGTCCTTTTGTGAGACTAAACCAATTATAAGTCTGGTGAGAAAATTAGAATATTTTGATGCACTACAGTACAGTTTAGAATATATAGAGTATTATCTGATTTAGCTTCCAGGGCAAAACATGTTTTAATCAGGTCTGCCAACCCAACAATATCTGTCGCTCATGAGCCCCATTAATTAATTAAGCACTTTAATTTCTAATTGACCGCTTTACGGTTTATAACATATATACGTCGTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/17/2015 at 03:57 AM