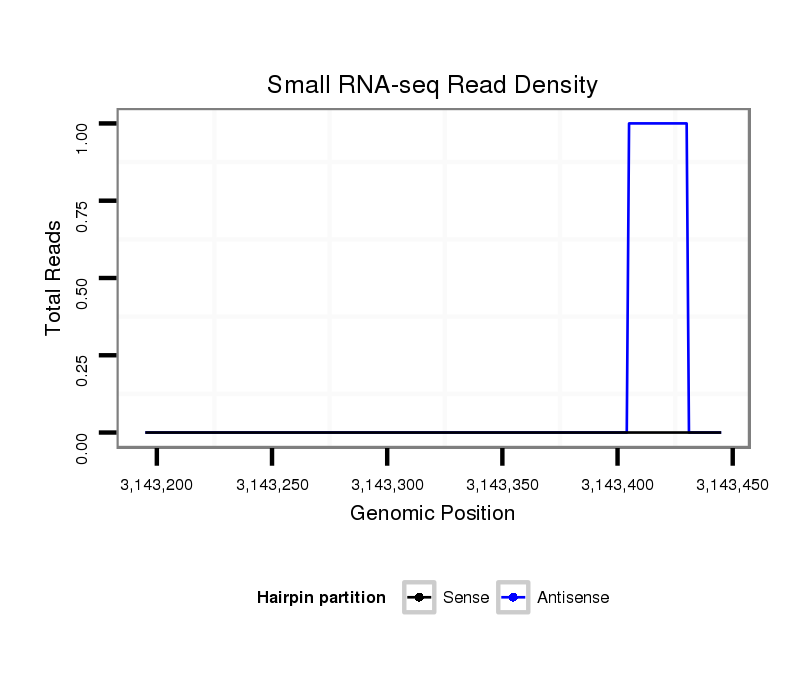
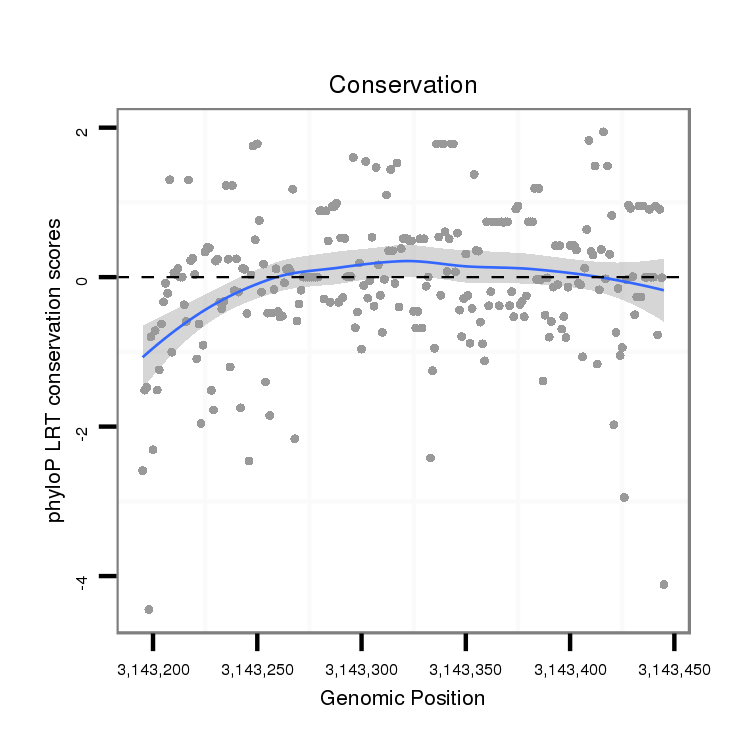
| AGCGCCGAGTCA--------------GCGCACCGATCAGCACCA------------------------CCGTGAGAGCCATC---GT-----------------GC-CACAAGACACCTAT-------C---AG------------CA---------------------GC---AA---CAGCAGTGACCACGGCGGCAG--------------------CGACGC-----CAATAGCAGCAG-----------CGGCAGCTTTC----------ATATAAATAGTACTTGTATAATTA-----------------TTCTTAAGTTAGTTCTAA-TCCAACTCCGA-------------AA-----------------------------------GTGAATAAATG-TTTTTTTAA--TCCAAAAACCGCTTTCGAATATTTAATTTATATATACTT-------------TGTGGGGTCTGCCACGCCTCCTT | Size | Hit Count | Total Norm | Total | M046
Female-body | V041
Embryo | V049
Head | V056
Head | M060
Embryo | V110
Male-body |
| .................................................................................................................ACACCTAT-------C---AG------------CA---------------------GC---AA---CAGCAGTGACC................................................................................................................................................................................................................................................................................................................ | 28 | 11 | 0.64 | 7 | 3 | 1 | 0 | 0 | 3 | 0 |
| .................................................................................................................ACACCTAT-------C---AG------------CA---------------------GC---AA---CAGC....................................................................................................................................................................................................................................................................................................................... | 21 | 12 | 0.58 | 7 | 2 | 0 | 0 | 0 | 5 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................TCCGA-------------AA-----------------------------------GTGAATAAATG-TTTTTTT............................................................................... | 25 | 10 | 0.50 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| ..........................................CA------------------------CCGTGAGAGCCATC---GT-----------------GC-CACAAG............................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................ACACCTAT-------C---AG------------CA---------------------GC---AA---CAGCA...................................................................................................................................................................................................................................................................................................................... | 22 | 12 | 0.33 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................ACACCTAT-------C---AG------------CA---------------------GC---AA---CAGCAG..................................................................................................................................................................................................................................................................................................................... | 23 | 12 | 0.25 | 3 | 2 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................ACACCTAT-------C---AG------------CA---------------------GC---AA---CAGCAGT.................................................................................................................................................................................................................................................................................................................... | 24 | 11 | 0.18 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................CACCTAT-------C---AG------------CA---------------------GC---AA---CAGCAGT.................................................................................................................................................................................................................................................................................................................... | 23 | 11 | 0.18 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................CACCTAT-------C---AG------------CA---------------------GC---AA---CAGCAG..................................................................................................................................................................................................................................................................................................................... | 22 | 12 | 0.17 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................ATAAATG-TTTTTTTAA--TCCAAAAACCGC............................................................... | 28 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................TCCAAAAACCGCTTTCGAATATTTA.................................................. | 25 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................TCCGA-------------AA-----------------------------------GTGAATAAATG-TTTTTTTA.............................................................................. | 26 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................TTTTTAA--TCCAAAAACCGCC.............................................................. | 20 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................CCGA-------------AA-----------------------------------GTGAATAAATG-TTTTTTT............................................................................... | 24 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................TCCGA-------------AA-----------------------------------GTGAATAAATG-TTTTTTTC.............................................................................. | 26 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................ACACCTAT-------C---AG------------CA---------------------GC---AA---CAGCAGTGAC................................................................................................................................................................................................................................................................................................................. | 27 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................ACACCTAT-------C---AG------------CA---------------------GC---AA---CAGCAGTGA.................................................................................................................................................................................................................................................................................................................. | 26 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................TCCGA-------------AA-----------------------------------GTGAATAAATG-TTTTTT................................................................................ | 24 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................ACCTAT-------C---AG------------CA---------------------GC---AA---CAGCAGTGACCAC.............................................................................................................................................................................................................................................................................................................. | 28 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................ACACCTAT-------C---AG------------CA---------------------GC---AA---CAGCAGTGACCA............................................................................................................................................................................................................................................................................................................... | 29 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................TCCGA-------------AA-----------------------------------GTGAATAAATG-TTTTT................................................................................. | 23 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................TTCTAA-TCCAACTCCGA-------------AA-----------------------------------GTGAAT............................................................................................ | 25 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................TCCAACTCCGA-------------AA-----------------------------------GTGAATAAATG-TTT................................................................................... | 27 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................TTCTAA-TCCAACTCCGA-------------AA-----------------------------------GTGA.............................................................................................. | 23 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................TCTTAAGTTAGTTCTAA-TCCAACTCCGA-------------....................................................................................................................................... | 28 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................TTA-----------------TTCTTAAGTTAGTTCTAA-TCCAAC......................................................................................................................................................... | 27 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................AGTTCTAA-TCCAACTCCGA-------------AA-----------------------------------GTGA.............................................................................................. | 25 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................AAGTTAGTTCTAA-TCCAACTCCGA-------------AA-----------------------------------.................................................................................................. | 26 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................................ATTA-----------------TTCTTAAGTTAGTTCTAA-TCC............................................................................................................................................................ | 25 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................G------------CA---------------------GC---AA---CAGCAGTGACCACGGC........................................................................................................................................................................................................................................................................................................... | 23 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................CGACGC-----CAATAGCAGCAG-----------CGG............................................................................................................................................................................................................................................. | 21 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................ATATATACTT-------------TGTGGGGTCTGC........... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |