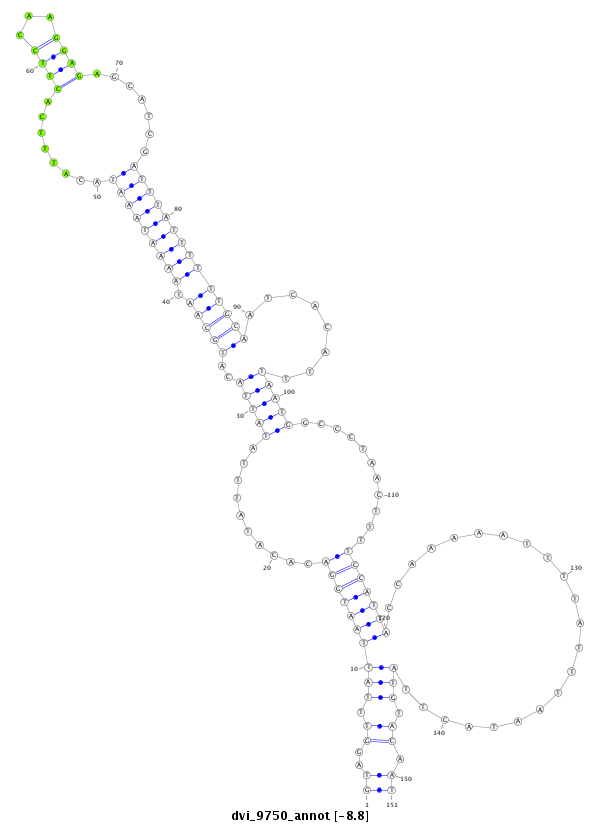
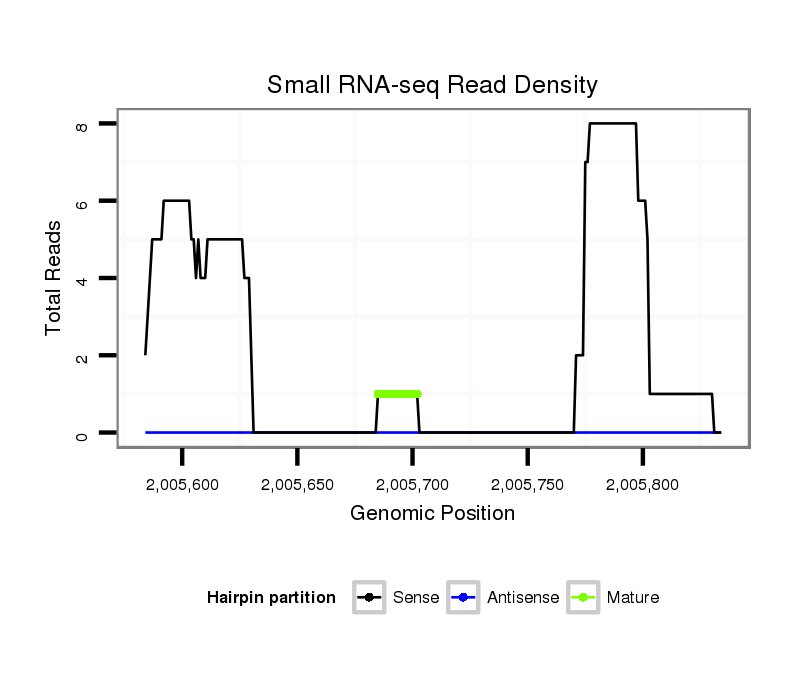
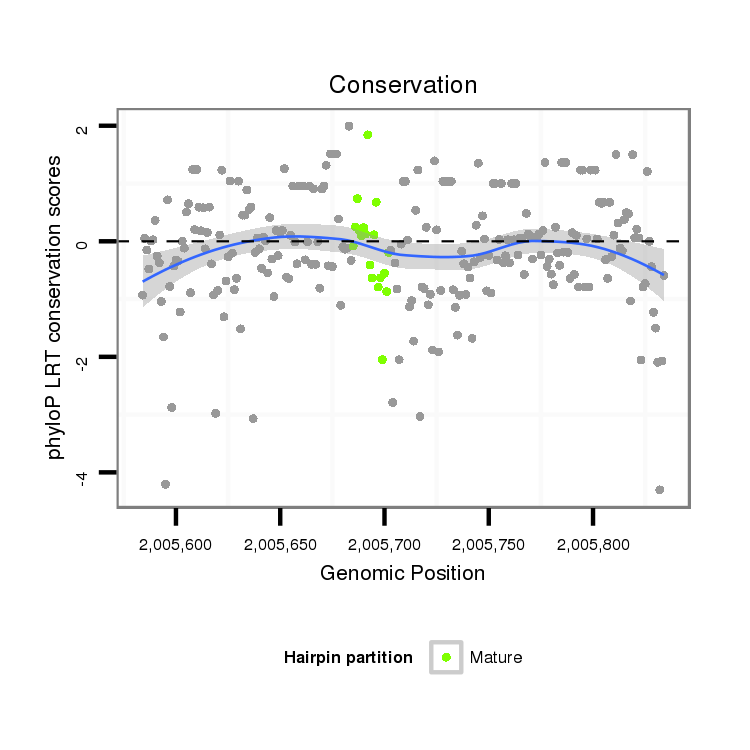
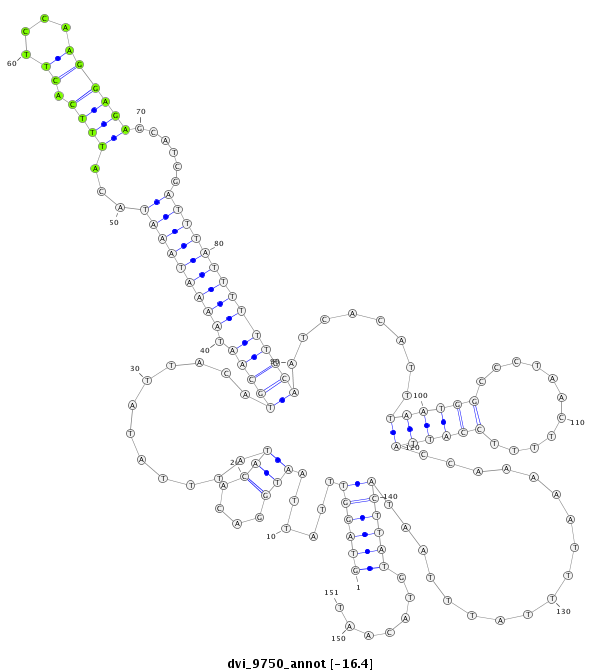
ID:dvi_9750 |
Coordinate:scaffold_12958:2005634-2005784 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -16.5 | -16.4 | -16.4 | -16.2 |
 |
 |
 |
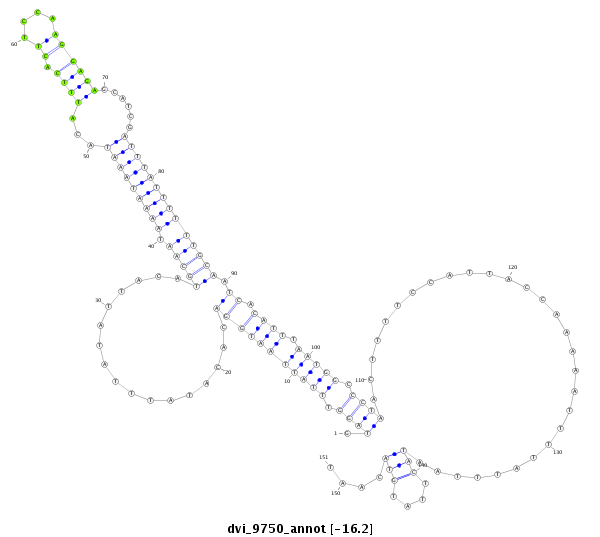 |
CDS [Dvir\GJ11187-cds]; exon [dvir_GLEANR_11177:2]; intron [Dvir\GJ11187-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GATATATTCCGTTGGGAAGTCCTTATAATTGGACCACCAGACACGCTTTAGTAGGTTTATTAATGGACACATATTTATATTACATGCAATAAAATAAATACATTTCACTTCCAAGGAGAGCATCGATTTATTTTTTGCAATCACATTTAATGGCCCTAACTTTTCCATTACCAAAAATTTTATTTAATACTTATGTACAATTGACTATAGTTCTTAATTTGATCACAAAATATAAAAAACATATAACTCCA **************************************************((..((.((((((((((..........(((((..(((((.(((((((((........((((...)))).......))))))))))))))........)))))...........)))))))......................))).)).))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
M047 female body |
SRR060689 160x9_testes_total |
M061 embryo |
SRR060658 140_ovaries_total |
SRR060659 Argentina_testes_total |
SRR060663 160_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060673 9_ovaries_total |
SRR060680 9xArg_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060675 140x9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................TATGTACAATTGACTATAGTTCTTAATT................................ | 28 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GATATATTCCGTTGGGAAGT....................................................................................................................................................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TACTTATGTACAATTGACTATAGTTCT..................................... | 27 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CCGTTGGGAAGTCCTTATAA............................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .ATATATTCCGTTGGGAAGTCCTT................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TATATTCCGTTGGGAAGTCCTTATAATC............................................................................................................................................................................................................................. | 28 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...ATATTCCGTTGGGAAGTCCTT................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................TTTTCCATTACCAAAAATTTTATTTAACC.............................................................. | 29 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................GACCACCAGACACGCTTTACGAA..................................................................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TGATCACAAAATATAAAAAACATATAAC.... | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TATATTCCGTTGGGAAGTCC..................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TCCGTTGGGAAGTCCTTCTAATTGGA.......................................................................................................................................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TGTACAATTGACTATAGTTCTTAATT................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................CCTTATAATTGGACCACCAGACACGCT............................................................................................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................ATTGGACCACCAGACACGC............................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................TATGTACAATTGACTATAGTTCTTAAT................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................ATTGGACCACCAGACACGCC............................................................................................................................................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................TTGGACCACCAGACACGCT............................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................TATAATTGGACCACCAGACACGC............................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................ATTTCACTTCCAAGGAGA.................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................ATAATTGGACCACCAGACA................................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................ATCACAGTTAAGGGACCTAA............................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................................TTTCGTCACAGAATATAAAAA............. | 21 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
|
CTATATAAGGCAACCCTTCAGGAATATTAACCTGGTGGTCTGTGCGAAATCATCCAAATAATTACCTGTGTATAAATATAATGTACGTTATTTTATTTATGTAAAGTGAAGGTTCCTCTCGTAGCTAAATAAAAAACGTTAGTGTAAATTACCGGGATTGAAAAGGTAATGGTTTTTAAAATAAATTATGAATACATGTTAACTGATATCAAGAATTAAACTAGTGTTTTATATTTTTTGTATATTGAGGT
**************************************************((..((.((((((((((..........(((((..(((((.(((((((((........((((...)))).......))))))))))))))........)))))...........)))))))......................))).)).))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060687 9_0-2h_embryos_total |
V116 male body |
SRR060682 9x140_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................TCCTCTCCTAGCTAA........................................................................................................................... | 15 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CTCGTAGGTAATTAAAAAA................................................................................................................... | 19 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................GGGATTGGAAAGCTAATGTT.............................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................AATGGTAATGGTTCTTGAAA...................................................................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................GGATTGAAAAGGTAAAGCC.............................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................GAAAAGGTACAGGTTTTTAG........................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12958:2005584-2005834 + | dvi_9750 | GAT-ATATTCCGTTGG--------G--------------------------------------------AA----GT-----------------------------------------------CCTT---------------------ATAATTGGAC-----------------------------CACCAGACACGCTTTAGTAGG--------------------------------TTTATTA------ATGGACACAT----------------------------------------------------------------------------------------------------------------ATTTATATTACATGCAATA--------AAATAAATACATTTCA------CTTCCAAGGAGAGCAT-C-----------------GAT----TT-----------------------ATTTTTTGCA-AT-CACATTTAATGGCCC-TAAC-TTTTCC------ATTACCAAAAATTTTATTTAATACTTATGTACAA---TTGACTA-TAGTTCTTAATTTGATCACAAAATAT-AAAA----AACATATAACTCCA-- |
| droMoj3 | scaffold_6498:2983312-2983538 - | AAT-GTACTATTTTTT--------G--------------------------------------------TT----AT-----------------------------------------------CTTA---------------------ATAAAACGATTATTACAGTTAGATGCTATATATAATTATATTCAAC----------CAGC---------------------TTTACA------------------------------------------------------------------------------------------------------------------------------------------------------------ACA--------AAATCGAAACCATTCACTTTTA------A-------------------------------ATTTAT--------ACTCTATGCATACTGTCTTAAAGAT-TT-----------------TCAT-TTTGCA------TTTATCATGATTTTTCACTAATCCTAATGCATTAACATTTACAATAGATTTTAAATT-------TACTCAA-ACAA----CAGG----AAAGAC-- | |
| droGri2 | scaffold_15116:363711-363871 - | GAT-ATATTCCGTTGG--------G--------------------------------------------AA----GT-----------------------------------------------CCTA---------------------ATAATTGGAC-----------------------------CACCAGATACACTGTAGTAGG--------------------------------TTTATTACAG-------------------------------------------------------------------------------T-AATTATATATATAT------------------------------ATATATGTTTCTTACAATACAATACAGCAATGAACACATTTCA------TTTCTAAGCATTAATTTTGCTCCC-----------------------------------------------------------------------------------------TCA----------------------------------------------------------------------A--T--TA-------GTTTCAA-- | |
| droWil2 | scf2_1100000004943:13816755-13816867 - | GAC-ATATTCCGGTGG--------G--------------------------------------------AA----GT-----------------------------------------------TCTC---------------------ATAATTGGGC-----------------------------CACCAGATACGCTTTAGTAAG--------------------------------TTCAGTGATC-------------------------------------------------------------------------------C-AATCACACA----------------------------------------------------------------------------------CA----------------------------------G---ATTT---------ATTTCTTTACCTATAGTGT-GAAAAT-----------------------------------------------------------------------------------------------------------------------------CTTTCA-- | |
| dp5 | 2:28151672-28151886 - | CTT-TATTATATATAT--------A--------------------------------------------AT----CTAAACATAAAAATACTGTAAAAT-----------------------------------------------------------------------------AAATATAAATATATTTAAT----------TAAA--------------------------------TAA------ATAATTAAATAATTAA----------------------------------------------------------------------------AAATAAAATATATTTCAATCAAATATTTTCAATT--------------------------------TCTACATTACT------TTTATATTTTTAAATTTCTTCTCT-------------------------------------------------------------------------------------------------------------AGAATTAATTCCTAA---ATAATTA-AATTTAAAAATCTGAAGATAAAATAT-ATAA----AAAA----TATATT-- | |
| droPer2 | scaffold_6:472308-472388 - | GAT-ATATTCCGCTGG--------G--------------------------------------------AA----GT-----------------------------------------------CCTA---------------------ATAATTGGTC-----------------------------CACCAGATACGCTTTAGTATG--------------------------------TCTAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAT-----------------------------------------------------------------------TTA--GATAT-------TTAATGTAA | |
| droAna3 | scaffold_13417:4944819-4944986 - | GAA-AAGTTCCATTGA--------C--------------------------------------------AA----GG----------ACTTTCAACAT---------------------------------------------------GCGACTAAAC-----------------------------TGCC----TTAAGGGGGCAGG--------------------TTGG--------TTTCCGA------GG--------CTAAAAAAAAAACAGATTTTTGTGATTTAAAAAAAAATTTATTTTTTTT-------------------------------------------ATTTTTTAATATAATT-------------------------------------------------------------ATTTAATTTTTTTTAC--------------------------------------------------------------------------------------------------------ACA----------------------------------------------------------------ATACATGGCTATA-- | |
| droBip1 | scf7180000395123:8168-8343 + | GACTATATCCCGTTTG--------A--------------------------------------------A------T-----------------------------------------------TTTG---------------------GGGGTCAAAT-----------------------------CCTCGACCAGCGTTCC-------------------------------GTTAACACCATTACAA----TAAATAAGTAT----------------------------------------------------------------------------AGCTAATATATACGTTAGTA---------------------------------------------TAGTATATTTTA------TTGCTACGTGAAACAC-CGGCCGCGGAA------------TT-AATTTTCCATTTCTTAACT---------------C--------------------------ATTTT----TAA----------------------------------------------------------------------A--A--AT-------GTCATTT-- | |
| droKik1 | scf7180000302577:1514300-1514440 - | GAA-AGATATTAACTA-----------------------------------------------------------TT-----------------------------------------------CTTA--------------A------ATAAATAAAT-----------------------------ATTCAAAAATTATTTAA----------------------------------------------------------------------------------------------------TTTTAATTTTTAATACCTAATATTATTT---------TTAATTACTTTAAACTATATATATATTTCAA---------------------------------------------------A----------------------------------A---ATTT---------ATTT----ATT---------------T--------------------------ATTTA----TAA----------------------------------------------------------------------A--A--AT-------ATTTAAT-- | |
| droFic1 | scf7180000453974:22317-22373 - | GAT-ATATTTCGGTGG--------G--------------------------------------------AA----GT-----------------------------------------------CTTA---------------------ATAATTGGAC-----------------------------CACCAGATACATTGTAGTATG--------------------------------TT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000490404:2919-2998 - | GAT-ATATTTCGCTGG--------G--------------------------------------------AA----GT-----------------------------------------------CTTA---------------------ATAATTGGAC-----------------------------CACCAGATACACTATAGTACG--------------------------------TT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT-TA------------------------------------ATAT-----------------------------------------------------------------------TTA--ATTAT-------TTACTTT-- | |
| droRho1 | scf7180000777990:14059-14160 + | AAT-TTATTAAAATGT--------A--------------------------------------------TAT---AA-----------------------------------------------TTAC---------------------ATAATTAGAT-----------------------------TGAGAAC----------TTGG--------------------------------TTTATAA------AA--------CT------------------------------------------------------------------AAAAA---------G-----------------------------AAATATAATACAACCAA--------------------------------------------------------------------------------------------------------------TTATA-------------A------------ATT--A---------------------------------------------------------------------T-A--A--AA-------TACATTT-- | |
| droBia1 | scf7180000299515:37328-37480 + | GAT-ATTTGTGGGTGCTAGAGTGGG--------------------------------------------CGT---GA-------------------------------------------------------CAAACTTTTTTT-------------------------------------------------------------TAGG--------------------------------TCA------ATCGATAGGTACTGAT--------------------------------------------------------------------------------------------------------------------------------A--------AAACTAATACATTTCA------CTTCTCAGGAAATC-C-CGGAATCTGCATGCCAAG---GCTG---------A--------------------------------------CTGCTC-TAGC-TTTTAC-ATTTTCC----------------------------------------------------------------------A--C--AT-------ATTTTAT-- | |
| droTak1 | scf7180000414562:243571-243774 + | ACT-GAAAGTCAT---------------------------------------------------------AC---AA-----------------------------------------------TTATAATT--GATACTTTTTGTTAA----------A------------------------------------------------------------------------------------------------------------------------TTTTCACATTTTTGAAAAAAGTTGATTTTTTTTTGTTACGCCCGACATCTC-G---------GAAATGCCCATATTTTTTTTAAACATTTTTAATT--------------------------------TTTACTT--TT------TTAATAATGTGAAATAAATGTATA-------------------------------------------------------------------------------------------------------------GGCACATTTTCTAAG---GTAACC----------------------AAAACT-GAAA----ATCATACAATTATA-- | |
| droEug1 | scf7180000408987:68796-69090 - | TTT-TTTTTTTAAAAAAAAAATGTGTGTAAAATTAAATTTTTTCTTAAAAAAAAATTTTTAGGTTTTTTTTTATTCTATTTATTAAAATTTTGTAAATTTTAGTAAATTAAAATTAGTGAAATTTTTT---------------------AAATTTGAAT-----------------------------TTTTAACAATTTTTTAGTAAAAATTTATTTTTTATTTTTAATTA----------TTG-------------------TA--------------------------------------------------------------------------------------------------------------------------------A--------TAATTAATAAATTTTA------TTTTTATATAATAATTAAGT-TTAA-----------AT----AT-----------------------TTTTATTATA--T-TTTATTTATTAATAA-ATTTATTTTTTA----TAA---------------------------------------------------------------AAATAT-TAAA-----ATCAATAATTAAT-- | |
| dm3 | chrM:16452-16646 + | TAT-TTATTAAATTAT--------A--------------------------------------------CTT---AA-------------------------------------------------------TAAACTATTT-------------------TTATAAT------------------------AAA----------TTAT-------TTTATAAATAAAATTATTTA------------------------------------------------------------------------------------------------------------------------------------------------------------AAA--------TAATTAATAAGAAATATTTTTATTATAATAAAAATTAA-A-----------------AAT----AA-----------------------TTTTTAAAAA-AT-TCAATTTATATTTAT-ATAT-ATA----------------------------------TATATATAT---ATAATTT-TAATTTTCAATT-------AAATTAT-ATAA----ATAT----AATAAA-- | |
| droSim2 | 2r:1013902-1013994 + | GAT-ATATTTCGCTGG--------G--------------------------------------------AA----GT-----------------------------------------------TTTA---------------------ATAATTGGAC-----------------------------CGCCAGATACACTGTAGTATG--------------------------------CT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT-GAAAAT-TTTTA-------------T------------TAT----TATTATATTTATGCAA-------------------------------------------------------------------------CAA-- | |
| droSec2 | scaffold_64:130833-130925 + | GAT-ATATTTCGCTGG--------G--------------------------------------------AA----GT-----------------------------------------------TTTA---------------------ATAATTGGAC-----------------------------CGCCAGATACACTGTAGTATG--------------------------------CT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT-GAAAAT-TTTTA-------------T------------TAT----TATTATATTTATGCAA-------------------------------------------------------------------------CAA-- | |
| droYak3 | v2_chrUh_007:47395-47524 - | ATC-ATAAAT-------------------------------------------------------------------------------------------------------------------------------ACTTTATGAGAT----------TATTTCAC--------------------------------------------------------------------ATTGATATTG-------------------TT--------------------------------------------------------------------------------------------------------------------------------A--------TAATAAAAATATTTTA------TTATAACAAAAAAGAA-T-----------------AAT----AC-----------------------GTCTCTTATA-CT-TTTATTTAATGGTAC-TTCT-TTTTTC------AT----------------------------------------------------------------------A--A--AC-------AAAGTTT-- | |
| droEre2 | scaffold_4784:23474939-23474995 + | GAT-ATATTTCGCTGG--------G--------------------------------------------AA----GT-----------------------------------------------TTTA---------------------ATAATTGGAC-----------------------------CGCCAGATACACTGTAGTATG--------------------------------TT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 11:06 PM