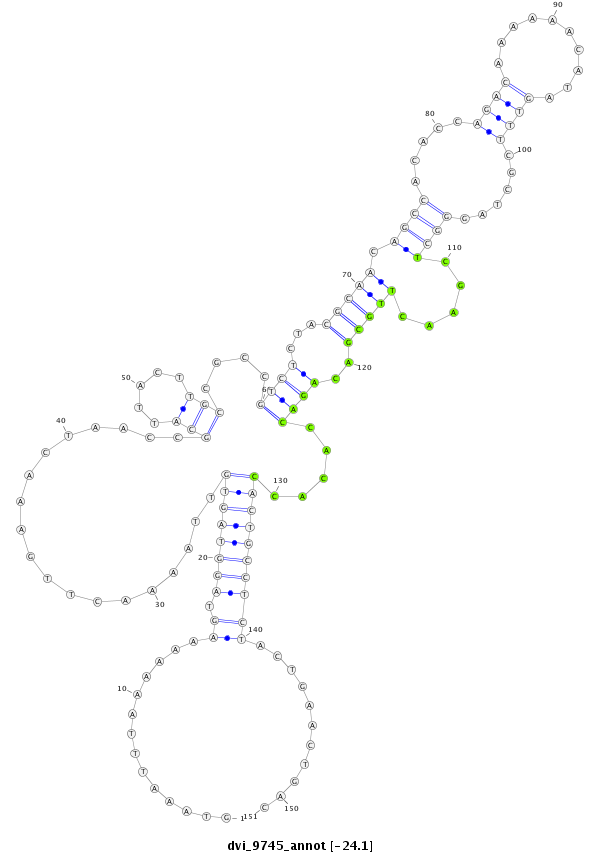
ID:dvi_9745 |
Coordinate:scaffold_12958:1909414-1909564 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -23.9 | -23.6 | -23.4 |
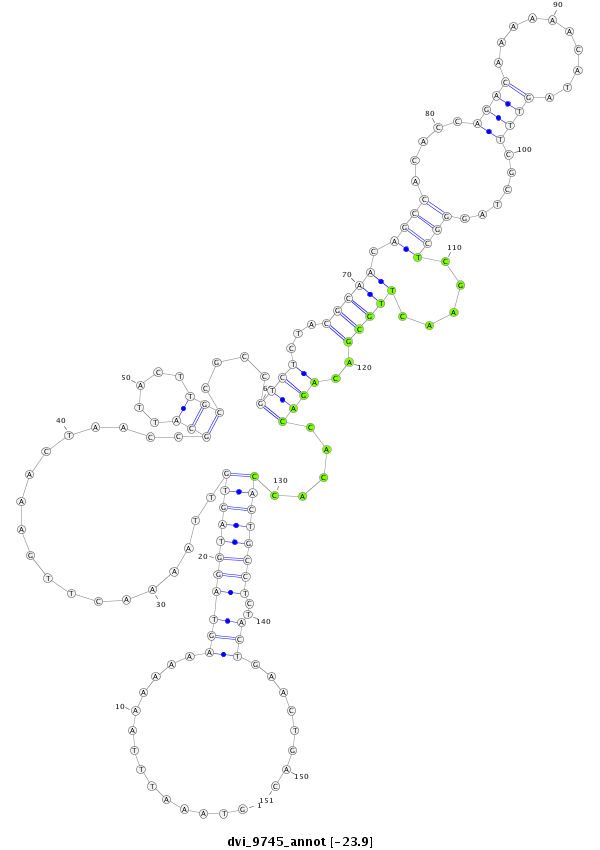 |
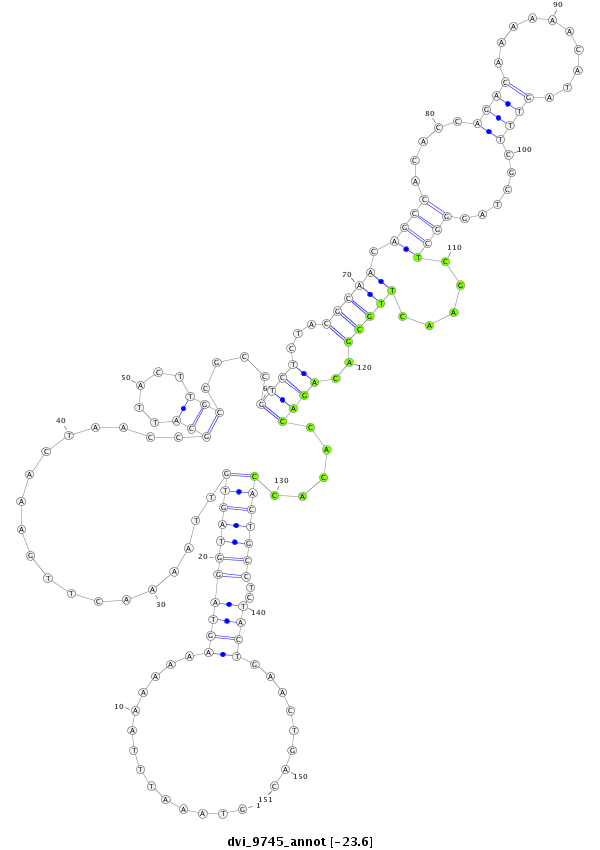 |
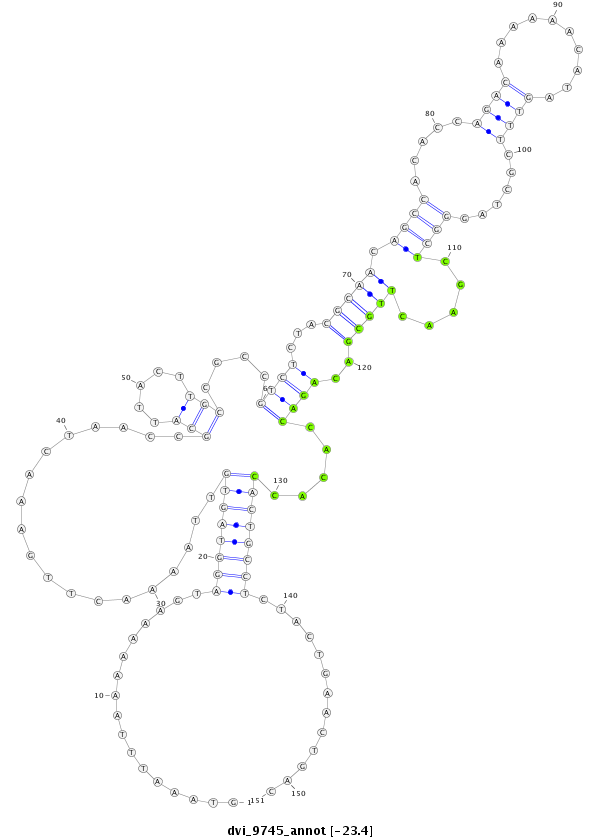 |
intron [Dvir\GJ11185-in]; exon [dvir_GLEANR_11173:2]; CDS [Dvir\GJ11185-cds]; intron [Dvir\GJ11185-in]
No Repeatable elements found
| mature | star |
| ------------######################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TTTCAACACCAGGTGCAACTCGCTCTACGTGCTCGTCTGGAGTAAAAAGGGTAAATTTAAAAAAAGTAGGTAGTGTTAAAACTTGAAACTAACCGCATTACTTGCCGCCGTCTCTACGCAACAGCCACACCAGACAAAAAACATAGTTTCGCTAGGGCTCGAACTTGCGACAGACCACACCACTGCCTCTACTGAACTGACATTTATGCGCGACTGCTTCGACTTCGATTGCATTGTTTTCTGTGCTGCAC **************************************************..............((.((((((((...................(((.....)))....((((...(((((.((((.....((((..........))))......)))).....)))))..)))).....))))))))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060677 Argx9_ovaries_total |
SRR060678 9x140_testes_total |
SRR060682 9x140_0-2h_embryos_total |
M028 head |
SRR060679 140x9_testes_total |
V047 embryo |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................ACATTTATGCGCGACTGCTTC............................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................TCGAACTTGCGACAGACCACACC...................................................................... | 23 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TATGTGCAACTGCTTCGA............................. | 18 | 2 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................ACTTGCTCGTCCGGTGTA............................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................CAACATTTATGCGCGACT.................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGCTTCAACTTCGTTTGCAC................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................GCCACACGAGACTACAAAC............................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
AAAGTTGTGGTCCACGTTGAGCGAGATGCACGAGCAGACCTCATTTTTCCCATTTAAATTTTTTTCATCCATCACAATTTTGAACTTTGATTGGCGTAATGAACGGCGGCAGAGATGCGTTGTCGGTGTGGTCTGTTTTTTGTATCAAAGCGATCCCGAGCTTGAACGCTGTCTGGTGTGGTGACGGAGATGACTTGACTGTAAATACGCGCTGACGAAGCTGAAGCTAACGTAACAAAAGACACGACGTG
**************************************************..............((.((((((((...................(((.....)))....((((...(((((.((((.....((((..........))))......)))).....)))))..)))).....))))))))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
M027 male body |
SRR060663 160_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
M047 female body |
SRR060669 160x9_females_carcasses_total |
V053 head |
SRR060662 9x160_0-2h_embryos_total |
SRR060655 9x160_testes_total |
GSM1528803 follicle cells |
SRR060664 9_males_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060681 Argx9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................TCCCGAGCTTGGACACTGG............................................................................... | 19 | 3 | 20 | 4.00 | 80 | 56 | 1 | 17 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................TCCCGAGCTTGGACACTG................................................................................ | 18 | 2 | 20 | 1.45 | 29 | 3 | 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................GCTAACGTACCAAAAGACAC...... | 20 | 1 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................ACCTCTTATTTCCCATTTTAAT................................................................................................................................................................................................ | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................GAAAGCGATGCCGAGGTTGAA..................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................CAGCGAGAGTCACGAGCAG...................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................CCCGAGCTTGAACGCTGTCTGG........................................................................... | 22 | 0 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................GCGCTGACGAACCTGATGT........................ | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TCCCGAGCTTGGACGCTG................................................................................ | 18 | 1 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TCCCGAGCTTGGATGCTG................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TCCCGAGCTTGGACACT................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................TCCCGAGCTTGGACACTGTG.............................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TCCCGAGCTTGGATGCTGGC.............................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................ATCCCGAGCTTGGATGCTG................................................................................ | 19 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GAAGCTGAAGCTACTGTAG................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................TCCCGAGCTTGGACGCTGG............................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12958:1909364-1909614 + | dvi_9745 | TTTCAACACCAGGTGCAACTCGCTCTACGTGCTCGTCTGGAGTAAAAAGGGTAAATTTAAAAAAAGTAGGTAGTGTTAAAACTTGAAACTAACCGCATTACTTGCCGCCGTCTCTACGCAACAGCCACACCAGACAAAAAACATAGTTTCGCTAGGGCTCGAACTTGCGACAGACCACACCACTGCCTCTACTGAACTGACATTTATGCGCGACTGCTTCGACTTCGATTGCATTGTTTTCTGTGCTGCAC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/16/2015 at 11:04 PM